Abstract
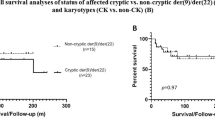
Deletions from the derivative chromosome 9, der(9), of the translocation, t(9;22)(q34;q11), at the site of the ABL/BCR fusion gene, have been demonstrated by fluorescence in situ hybridisation (FISH), in both Philadelphia chromosome (Ph)-positive chronic myeloid leukaemia (CML) and acute lymphoblastic leukaemia (ALL). In CML they occur in 10–15% of cases and appear to indicate a worse prognosis, whereas in ALL, the situation is unclear. This study presents the findings of dual fusion FISH used to detect such deletions in a series of 27 BCR/ABL-positive childhood ALL patients. Metaphase FISH was essential for the accurate interpretation of interphase FISH signal patterns. Three cases (11%) had a single fusion signal, resulting from deletions of the der(9). Three other patients with variant translocations and one with an insertion, also had a single fusion, but with no evidence of deletions. Gain of a fusion in approximately one-third of patients indicated a second Ph, which appears to be a diagnostic marker of Ph-positive ALL. This study shows that the incidence of deletions from the der(9) in childhood ALL is at least as high as that reported for CML.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Secker-Walker LM . Chromosomes and Genes in Acute Lymphoblastic Leukaemia. New York: Chapman and Hall, 1997.
Pui CH, Evans WE . Acute lymphoblastic leukemia. N Engl J Med 1998; 339: 605–615.
Secker-Walker LM, Prentice HG, Durrant J, Richards S, Hall E, Harrison G . Cytogenetics adds independent prognostic information in adults with acute lymphoblastic leukaemia on MRC trial UKALL XA. Br J Haematol 1997; 96: 601–610.
Hann I, Vora A, Harrison G, Harrison C, Martineau M, Moorman AV et al. Determinants of outcome after intensified therapy of childhood lymphoblastic leukaemia: results from Medical Research Council United Kingdom acute lymphoblastic leukaemia XI protocol. Br J Haematol 2001; 113: 103–114.
Deininger MW, Goldman JM, Melo JV . The molecular biology of chronic myeloid leukemia. Blood 2000; 96: 3343–3356.
Sinclair PB, Nacheva EP, Leversha M, Telford N, Chang J, Reid A et al. Large deletions at the t(9;22) breakpoint are common and may identify a poor-prognosis subgroup of patients with chronic myeloid leukemia. Blood 2000; 95: 738–743.
Dewald GW, Wyatt WA, Juneau AL, Carlson RO, Zinsmeister AR, Jalal SM et al. Highly sensitive fluorescence in situ hybridization method to detect double BCR/ABL fusion and monitor response to therapy in chronic myeloid leukemia. Blood 1998; 91: 3357–3365.
Huntly BJ, Bench AJ, Delabesse E, Reid AG, Li J, Scott MA et al. Derivative chromosome 9 deletions in chronic myeloid leukemia: poor prognosis is not associated with loss of ABL-BCR expression, elevated BCR-ABL levels, or karyotypic instability. Blood 2002; 99: 4547–4553.
Huntly BJ, Bench A, Green AR . Double jeopardy from a single translocation: deletions of the derivative chromosome 9 in chronic myeloid leukemia. Blood 2003; 102: 1160–1168.
Huntly BJ, Reid AG, Bench AJ, Campbell LJ, Telford N, Shepherd P et al. Deletions of the derivative chromosome 9 occur at the time of the Philadelphia translocation and provide a powerful and independent prognostic indicator in chronic myeloid leukemia. Blood 2001; 98: 1732–1738.
Kolomietz E, Al Maghrabi J, Brennan S, Karaskova J, Minkin S, Lipton J et al. Primary chromosomal rearrangements of leukemia are frequently accompanied by extensive submicroscopic deletions and may lead to altered prognosis. Blood 2001; 97: 3581–3588.
Cohen N, Rozenfeld-Granot G, Hardan I, Brok-Simoni F, Amariglio N, Rechavi G et al. Subgroup of patients with Philadelphia-positive chronic myelogenous leukemia characterized by a deletion of 9q proximal to ABL gene: expression profiling, resistance to interferon therapy, and poor prognosis. Cancer Genet Cytogenet 2001; 128: 114–119.
Specchia G, Albano F, Anelli L, Storlazzi CT, Zagaria A, Mancini M et al. Deletions on der(9) chromosome in adult Ph-positive acute lymphoblastic leukemia occur with a frequency similar to that observed in chronic myeloid leukemia. Leukemia 2003; 17: 528–531.
Reid A, Huntly BJ, Henning E, Niederwiser D, Campbell LJ, Brown N et al. Deletions of the derivative chromosome 9 are rare in Philadelphia-positive acute lymphobalstic leukemia. Blood 2002; 99: 2274–2275.
Harrison CJ, Martineau M, Secker-Walker LM . The Leukaemia Research Fund/United Kingdom Cancer Cytogenetics Group Karyotype Database in acute lymphoblastic leukaemia: a valuable resource for patient management. Br J Haematol 2001; 113: 3–10.
ISCN. An International System for Human Cytogenetic Nomenclature (1995). 1st edn. Basel: S.Karger, 1995.
Schenk TM, Keyhani A, Bottcher S, Kliche KO, Goodacre A, Guo JQ et al. Multilineage involvement of Philadelphia chromosome positive acute lymphoblastic leukemia. Leukemia 1998; 12: 666–674.
Primo D, Tabernero MD, Rasillo A, Sayagues JM, Espinosa AB, Chillon MC et al. Patterns of BCR/ABL gene rearrangements by interphase fluorescence in situ hybridization (FISH) in BCR/ABL+ leukemias: incidence and underlying genetic abnormalities. Leukemia 2003; 17: 1124–1129.
Jabbar Al-Obaidi MS, Martineau M, Bennett CF, Franklin IM, Goldstone AH, Harewood L et al. ETV6/AML1 fusion by FISH in adult acute lymphoblastic leukemia. Leukemia 2002; 16: 669–674.
Storlazzi CT, Specchia G, Anelli L, Albano F, Pastore D, Zagaria A et al. Breakpoint characterization of der(9) deletions in chronic myeloid leukemia patients. Genes Chromosomes Cancer 2002; 35: 271–276.
Harewood L, Robinson H, Harris R, Jabbar Al-Obaidi MS, Jalali GR, Martineau M et al. Amplification of AML1 on a duplicated chromosome 21 in aute lymphoblastic leukemia: a study of 20 cases. Leukemia 2003; 17: 547–553.
Grand F, Kulkarni S, Chase A, Goldman JM, Gordon M, Cross NC . Frequent deletion of hSNF5/INI1, a component of the SWI/SNF complex, in chronic myeloid leukemia. Cancer Res 1999; 59: 3870–3874.
Reid AG, Huntly BJ, Grace C, Green AR, Nacheva EP . Survival implications of molecular heterogeneity in variant Philadelphia-positive chronic myeloid leukaemia. Br J Haematol 2003; 121: 419–427.
Primo D, Tabernero MD, Espinosa AB, Chillon MC, Gutierrez N, Orfao A . Reply to Wan et al: atypical fluorescence in situ hybridisation pattern in chronic myeloid leukaemia due to cryptic insertion of BCR at 9q34. Leukemia 2004; 18: 162–164.
Morel F, Herry A, Le Bris MJ, Morice P, Bouquard P, Abgrall JF et al. Contribution of fluorescence in situ hybridization analyses to the characterization of masked and complex Philadelphia chromosome translocations in chronic myelocytic leukemia. Cancer Genet Cytogenet 2003; 147: 115–120.
Reddy KS, Sulcova V . A FISH study of variant Philadelphia rearrangements. Cancer Genet Cytogenet 2000; 118: 121–131.
Loncarevic IF, Romer J, Starke H, Heller A, Bleck C, Ziegler M et al. Heterogenic molecular basis for loss of ABL1-BCR transcription: deletions in der(9)t(9;22) and variants of standard t(9;22) in BCR-ABL1-positive chronic myeloid leukemia. Genes Chromosomes Cancer 2002; 34: 193–200.
Wan TS, Ma SK, Li CK, Chan LC . Atypical fluorescence in situ hybridisation pattern in chronic myeloid leukaemia due to cryptic insertion of BCR at 9q34. Leukemia 2004; 18: 161–162.
Heerema NA, Harbott J, Galimberti S, Camitta BM, Gaynon PS, Janka-Schaub G et al. Secondary cytogenetic aberrations in childhood Philadelphia chromosome positive acute lymphoblastic leukemia are nonrandom and may be associated with outcome. Leukemia 2004; 18: 693–702.
Herens C, Tassin F, Lemaire V, Beguin Y, Collard E, Lampertz S et al. Deletion of the 5′-ABL region: a recurrent anomaly detected by fluorescence in situ hybridization in about 10% of Philadelphia-positive chronic myeloid leukaemia patients. Br J Haematol 2000; 110: 214–216.
Lee DS, Lee Y, Yun Y, Kim Y, Jeong SS, Lee YK et al. A study on the incidence of ABL gene deletion on derivative chromosome 9 in chronic myelogenous leukemia by interphase fluorescence in situ hybridization and its association with disease progression. Genes Chromosomes Cancer 2003; 37: 291–299.
Kolomietz E, Marrano P, Yee K, Thai B, Braude I, Kolomietz A et al. Quantitative PCR identifies a minimal deleted region of 120 kb extending from the Philadelphia chromosome ABL translocation breakpoint in chronic myeloid leukemia with poor outcome. Leukemia 2003; 17: 1313–1323.
Reid AG, Nacheva EP . A potential role for PRDM12 in the pathogenesis of chronic myeloid leukaemia with derivative chromosome 9 deletion. Leukemia 2004; 18: 178–180.
Wan TS, Ma SK, Au WY, Chan LC . Derivative chromosome 9 deletions in chronic myeloid leukaemia: interpretation of atypical D-FISH pattern. J Clin Pathol 2003; 56: 471–474.
Acknowledgements
The authors would like to thank the Leukaemia Research Fund for financial support; Dr Mariano Rocchi, Italy, for PAC probes dJ1132H12 and dJ835J22; Dr John Crolla and his team, Wessex Regional Genetics Laboratory, Salisbury, for growing and preparation of the PAC probes; the Clinical Trial Service Unit (CTSU), Oxford, for clinical and survival data. The authors are also grateful to the following cytogenetics laboratories for providing cytogenetic and FISH data as well as fixed cell suspensions on this patient series: Academic Haematology and Cytogenetics, Royal Marsden NHS Trust, Surry; West Midlands Regional Genetics Services, Birmingham Women's Hospital; Leukaemia Cytogenetics, Addenbrooke's Hospital, Cambridge; National Centre for Medical Genetics, Our Lady's Hospital for Sick Children, Dublin; Duncan Guthrie Institute of Medical Genetics, Yorkhill NHS Trust Hospitals, Glasgow; Haematology Cytogenetic Laboratory, S.E Scotland Cytogenetic Services, Western General Hospital, Edinburgh; LRF Centre for Childhood Leukemia, Great Ormond Street Hospital, London; Oxford Medical Genetics Laboratories, Churchill Hospital, Oxford; Oncology Cytogenetics Service, Christie Hospital NHS Trust, Manchester; North Trent Cytogenetics Service, Sheffield Children's Hospital NHS Trust, Sheffield; Haematology Department, University College Hospital, London. This study could not have been performed without the dedication of the NCRI Childhood and Adult Leukaemia Working Parties and their members, who have designed and coordinated the clinical trials through which these patients were identified and through which they were treated.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Robinson, H., Martineau, M., Harris, R. et al. Derivative chromosome 9 deletions are a significant feature of childhood Philadelphia chromosome positive acute lymphoblastic leukaemia. Leukemia 19, 564–571 (2005). https://doi.org/10.1038/sj.leu.2403629
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.leu.2403629
Keywords
This article is cited by
-
A protease-resistant Escherichia coli asparaginase with outstanding stability and enhanced anti-leukaemic activity in vitro
Scientific Reports (2017)
-
An unusual case of high hyperdiploid childhood ALL with cryptic BCR/ABL1 rearrangement
Molecular Cytogenetics (2014)
-
Disruption of ETV6 in intron 2 results in upregulatory and insertional events in childhood acute lymphoblastic leukaemia
Leukemia (2008)