Abstract
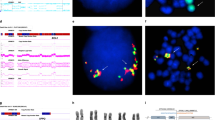
Heterozygous and homozygous deletions of chromosome 13q14.3 are found in 50% of patients with B cell CLL, suggesting the presence of one or more tumour suppressor genes within the deleted region. To identify candidate genes from the region, we constructed a map of 13q14.3 using a combination of genomic and cDNA library screening. The incidence of deletions in CLL patients was 51.5% encompassing a 265 kb region of minimal deletion (RMD) telomeric to markers D13S319. Two CpG islands were identified within the RMD, the telomeric of which is fully methylated whilst the more centromeric is unmethylated. A novel transcript was identified within the RMD that represents an alternative splice version of Leu1. The nine exons of this transcript span a genomic of 436 kb with exon 1 of Leu1 being the common first exon. The remaining exons were shown to be more frequently deleted than Leu1 itself. All splice forms of this transcript were detectable by RT-PCR but Leu1 detected the most abundant message on Northern blotting. Sequence analysis failed to reveal inactivating mutations in patients with heterozygous deletion of 13q14.3, although a polymorphic T to A variant was identified within exon 1 of Leu1 in leukemic and normal controls. As no mutations have been detected for Leu1 or any other transcript so far described, we cannot exclude the existence of control elements within the RMD that may regulate expression of genes lying in this region.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Corcoran MM, Rasool O, Liu Y, Iyengar A, Grander D, Ibbotson RE, Merup M, Wu X, Brodyansky V, Gardiner AC, Juliusson G, Chapman RM, Ivanova G, Tiller M, Gahrton G, Yankovsky N, Zabarovsky E, Oscier DG, Einhorn S . Detailed molecular delineation of 13q14.3 loss in B-cell chronic lymphocytic leukemia Blood 1998 91: 1382–1390
Panayiotidis P, Ganeshaguru K, Hoffbrand AV, Rowntree C, Jabbar SAB, Foroni L . Deletion of 13q14.3 and not 13q12 is the most common genetic abnormality detected in chronic lymphocytic leukemia cells Blood 1997 89: 734–735
Stilgenbauer S, Nickolenko J, Wilhelm J, Wolf S, Weitz S, Dohner K, Boehm T, Dohner H, Lichter P . Expressed sequences as candidates for a novel tumor suppressor gene at band 13q14 in B-cell chronic lymphocytic leukemia and mantle cell lymphoma Oncogene 1998 16: 1891–1897
Cave H, Avet-Loiseau H, Devaux I, Rondeau G, Boutard P, Lebrun E, Mechinaud F, Vilmer E, Grandchamp B . Deletion of chromosomal region 13q14.3 in childhood acute lymphoblastic leukemia Leukemia 2001 15: 371–376
Konigsberg R, Ackermann J, Kaufmann H, Zojer N, Urbauer E, Kromer E, Jager U, Gisslinger H, Schreiber S, Heinz R, Ludwig H, Huber H, Drach J . Deletions of chromosome 13q in monoclonal gammopathy of undetermined significance Leukemia 2000 14: 1975–1979
Lens D, Matutes E, Catovsky D, Coignet LJ . Frequent deletions at 11q23 and 13q14 in B cell prolymphocytic leukemia (B-PLL) Leukemia 2000 14: 427–430
Tanaka K, Arif M, Eguchi M, Guo SX, Hayashi Y, Asaoku H, Kyo T, Dohy H, Kamada N . Frequent allelic loss of the RB, D13S319 and D13S25 locus in myeloid malignancies with deletion/translocation at 13q14 of chromosome 13, but not in lymphoid malignancies Leukemia 1999 13: 1367–1373
Wada M, Okamura T, Okada M, Teramura M, Masuda M, Motoji T, Mizoguchi H . Frequent chromosome arm 13q deletion in aggressive non-Hodgkin's lymphoma Leukemia 1999 13: 792–798
Chang H, Bouman D, Boerkoel CF, Stewart AK, Squire JA . Frequent monoallelic loss of D13S319 in multiple myeloma patients shown by interphase fluorescence in situ hybridization Leukemia 1999 13: 105–109
Merup M, Jansson M, Corcoran M, Liu Y, Wu X, Rasool O, Stellan B, Hermansson M, Juliusson G, Gahrton G, Einhorn S . A FISH cosmid ‘cocktail’ for detection of 13q deletions in chronic lymphocytic leukemia – comparison with cytogenetics and Southern hybridization Leukemia 1998 12: 705–709
Navarro B, Garcia-Marco JA, Jones D, Price CM, Catovsky D . Association and clonal distribution of trisomy 12 and 13q14 deletions in chronic lymphocytic leukaemia Br J Haematol 1998 102: 1330–1334
Kalachikov S, Migliazza A, Cayanis E, Fracchiolla NS, Bonaldo MF, Lawton L, Jelenc P, Ye X, Qu X, Chien M, Hauptschein R, Gaidano G, Vitolo U, Saglio G, Resegotti L, Brodjansky V, Yankovsky N, Zhang P, Soares MB, Russo J, Edelman IS, Efstratiadis A, Dalla-Favera R, Fischer SG . Cloning and gene mapping of the chromosome 13q14 region deleted in chronic lymphocytic leukemia Genomics 1997 42: 369–377
Bouyge-Moreau I, Rondeau G, Avet-Loiseau H, Andre MT, Bezieau S, Cherel M, Saleun S, Cadoret E, Shaikh T, De Angelis MM, Arcot S, Batzer M, Moisan JP, Devilder MC . Construction of a 780-kb PAC, BAC, and cosmid contig encompassing the minimal critical deletion involved in B cell chronic lymphocytic leukemia at 13q14.3 Genomics 1997 46: 183–190
Bullrich F, Veronese ML, Kitada S, Jurlander J, Caligiuri MA, Reed JC, Croce CM . Minimal region of loss at 13q14 in B-cell chronic lymphocytic leukemia Blood 1996 88: 3109–3115
Liu Y, Corcoran M, Rasool O, Ivanova G, Ibbotson R, Grander D, Iyengar A, Baranova A, Kashuba V, Merup M, Wu X, Gardiner A, Mullenbach R, Poltaraus A, Hultstrom AL, Juliusson G, Chapman R, Tiller M, Cotter F, Gahrton G, Yankovsky N, Zabarovsky E, Einhorn S, Oscier D . Cloning of two candidate tumour suppressor genes within a 10 kb region on chromosome 13q14, frequently deleted in chronic lymphocytic leukemia Oncogene 1997 15: 2463–2473
Migliazza A, Bosch F, Komatsu H, Cayanis E, Martinotti S, Toniato E, Guccione E, Qu X, Chien M, Murty VV, Gaidano G, Inghirami G, Zhang P, Fischer S, Kalachikov SM, Russo J, Edelman I, Efstratiadis A, Dalla-Favera R . Nucleotide sequences, transcription map, and mutations analysis of the 13q14 chromosomal region deleted in B-cell chronic lymphocytic leukemia Blood 2001 97: 2098–2104
Rondeau G, Moreau I, Bezieau S, Cadoret E, Moisan JP, Devilder MC . Exclusion of Leu1 and Leu2 genes as tumor suppressor genes in 13q14.3-deleted B-CLL Leukemia 1999 13: 1630–1632
Kapanadze B, Kashuba V, Baranova A, Rasool O, van Everdink W, Liu Y, Syomov A, Corcoran M, Poltaraus A, Brodyansky V, Syomova N, Kazakov A, Ibbotson R, van den Berg A, Gizatullin R, Fedorova L, Sulimova G, Zelenin A, Deaven L, Lehrach H, Grander D, Buys C, Oscier D, Zabarovsky ER, Einhorn S, Yankovsky N . A cosmid and cDNA fine physical map of a human chromosome 13q14 region frequently lost in B-cell chronic lymphocytic leukemia and identification of a new putative tumour suppressor gene, Leu 5 FEBS Lett 1998 426: 266–270
Nehls M, Pfeifer D, Boehm T . Exon amplification from complete libraries of genomic DNA using a novel phage vector with automatic plasmid excision facility: Application to the mouse neurofibromatosis-1 locus Oncogene 1994 9: 2169–2175
Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, Sultan C . The French–American–British Co-operative Group: proposals for the classification of chronic (mature) B and T chronic lymphocytic leukaemias J Clin Pathol 1989 42: 567–584
Jabbar SAB, Ganeshaguru K, Wickremasinghe RG, Hoffbrand AV, Foroni L . Deletion of chromosome 13 (band q14) but not trisomy 12 is a clonal event in B-chronic lymphocytic leukaemia Br J Haematol 1995 90: 476–478
Liu Y, Szekely L, Grander D, Soderhall S, Juliusson G, Gahrton G, Linder S, Einhorn S . Chronic lymphocytic leukemia cells with allelic deletions at 13q14 commonly have one intact RB1 gene: evidence for a role of an adjacent locus Proc Natl Acad Sci USA 1993 90: 8697–8701
Baer R, Boehm T, Spits L, Rabbitts TH . Complex rearrangements in human J delta–C delta/J alpha–C alpha locus and aberrant recombination between J alpha segments EMBO J 1988 7: 1661–1668
Sanger F, Nicklen S, Coulsen AR . DNA sequencing with chain terminating inhibitors Proc Natl Acad Sci USA 1977 74: 5463–5467
Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, Smink LJ, Ainscough R, Almeida JP, Babbage A, Bagguley C, Bailey J, Barlow K, Bates KN, Beasley O, Bird CP, Blakey S, Bridgeman AM, Buck D, Burgess J, Burrill WD, O'Brien KP other 50 authors. The DNA sequence of human chromosome 22 Nature 1999 402: 489–495
Gyapay G, Morissette J, Vignal A, Dib C, Fizames C, Millasseau P, Marc S, Bernardi G, Lathrop M, Weissenbach J . The 1993–1994 Genethon human genetic linkage map Nat Genet 1994 7: 246–339
Jackson A, Carrara P, Duke V, Sinclair P, Papaioannou M, Harrison CJ, Foroni L . Deletion of 6q16–q21 in human lymphoid malignancies: a mapping and deletion analysis Cancer Res 2000 60: 2775–2779
Schwartz DC, Cantor CR . Separation of yeast chromosome-sized DNAs by pulsed field gel electrophoresis Cell 1984 37: 67–75
Sambrook J, Fritsch EF, Maniatis T . Molecular Cloning: A Laboratory Manual Cold Spring Harbour Laboratory Press: Cold Spring Harbor 1989
Jackson A, Panayiotidis P, Foroni L . The human homologue of the Drosophila tailless gene (TLX): characterization and mapping to a region of common deletion in human lymphoid leukemia on chromosome 6q21 Genomics 1998 50: 34–43
Chomczynski P, Sacchi N . Single step method of RNA isolation by acid guanidinum thiocyanate–phenol–chloroform extraction Anal Biochem 1987 162: 156–159
Rogan DF, Cousins DJ, Stanov DZ . Intergenic transcription occurs throughout the human IL-4/IL-13 gene cluster Biochem Biophys Res Commun 1999 255: 556–561
Knudson AG . Mutation and cancer: statistical study of retinoblastoma Proc Natl Acad Sci USA 1971 68: 820–823
Yoshikawa H, Matsubara K, Qian GS, Jackson P, Groopman JD, Manning JE, Harris CC, Herman JG . SOCS-1, a negative regulator of JAK/STAT pathway, is silenced by methylation in human hepatocellular carcinoma and shows growth-suppressor activity Nat Genet 2001 28: 29–35
Song WJ, Sullivan MG, Legare RD, Hutchings S, Tan X, Kufrin D, Ratajczak J, Resende IC, Haworth C, Hock R, Loh M, Felix C, Roy DC, Busque L, Kurnit D, Willman C, Gewirtz AM, Speck NA, Bushweller JH, Li FP, Gardiner K, Poncz M, Maris JM, Gilliland DG . Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia Nat Genet 1999 23: 166–175
Cai Z, de Bruijn M, Ma X, Dortland B, Luteijn T, Downing RJ, Dzierzak E . Haploinsufficiency of AML1 affects the temporal and spatial generation of hematopoietic stem cells in the mouse embryo Immunity 2000 13: 423–431
Fini ME, Bendena WG, Pardue ML . Unusual behaviour of the cytoplasmic transcript of hsr omega: an abundant, stress-inducible RNA that is translated but yields no detectable protein product J Cell Biol 1989 108: 2045–2057
Hao Y, Crenshaw T, Moulton T, Newcomb E, Tycko B . Tumour-suppressor activity of H19 RNA Nature 1993 365: 764–767
Barbour VM, Tufarelli C, Sharpe JA, Smith ZE, Ayyub H, Heinlein CA, Sloane-Stanley J, Indrak K, Wood WG, Higgs DR . Alpha-thalassaemia resulting from a negative chromosomal position effect Blood 2000 96: 800–807
Mabuchi H, Fujii H, Calin G, Alder H, Negrini M, Rassenti L, Kipps TJ, Bullrich F, Croce CM . Cloning and characterization of CLLD6, CLLD7, and CLLD8, novel candidate genes for leukemogenesis at chromosome 13q14, a region commonly deleted in B-cell chronic lymphocytic leukemia Cancer Res 2001 61: 2870–2877
Wolf S, Mertens D, Schaffner C, Korz C, Dohner H, Stilgenbauer S, Lichter P . B-cell neoplasia associated gene with multiple splicing (BCMS): the candidate B-CLL gene on 13q14 comprises more than 560 kb covering all critical regions Hum Mol Genet 2001 10: 1275–1285
Bullrich F, Fujii H, Calin G, Mabuchi H, Negrini M, Pekarsky Y, Rassenti Laura, Alder H, Reed JC, Keating MJ, Kipps TJ, Croce C . Characterisation of the 13q14 tumour suppressor locus in CLL: identification of ALT1, an alternative splice variant of the Leu2 gene Cancer Res 2001 61: 6640–6648
Acknowledgements
We would like to thank Dr Atul Mehta, MD, Professor HG Prentice, K Ganeshaguru, PhD and Steve Hart, PhD (Department of Hematology, Royal Free and University College School of Medicine) for assisting in the collection of material from CLL patients; Professor P Sfikakis (University of Athens, Laikon Hospital, Athens Greece) for financial support to Dr Panayiotidis and Dr Kotsi; Ann Walker, PhD (Department of Medicine, Royal Free and University College School of Medicine) for critically reviewing the manuscript and providing DNA from normal controls, and Andrew Dunham, PhD (The Sanger Centre, Hinxton, Cambs, UK) for invaluable help with the sequencing of BAC clones and gene analysis of chromosome 13. Research grant support: Kay Kendall Leukaemia Fund.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Rowntree, C., Duke, V., Panayiotidis, P. et al. Deletion analysis of chromosome 13q14.3 and characterisation of an alternative splice form of LEU1 in B cell chronic lymphocytic leukemia. Leukemia 16, 1267–1275 (2002). https://doi.org/10.1038/sj.leu.2402551
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.leu.2402551
Keywords
This article is cited by
-
DLEU1 promotes cell survival by preventing DYNLL1 degradation in esophageal squamous cell carcinoma
Journal of Translational Medicine (2022)
-
A study of Kibbutzim in Israel reveals risk factors for cardiometabolic traits and subtle population structure
European Journal of Human Genetics (2018)
-
Pro-apoptotic and antiproliferative activity of human KCNRG, a putative tumor suppressor in 13q14 region
Tumor Biology (2010)