Abstract
PKM2 (pyruvate kinase M2), a critical regulator of glycolysis, is phosphorylated by numerous growth factor receptors and oncogenic tyrosine kinases including NPM-ALK which is expressed in a subset of aggressive T-cell non-Hodgkin lymphomas known as anaplastic large cell lymphoma, ALK-positive. Our previous work demonstrated that phosphorylation of Y105-PKM2 by NPM-ALK regulates a major metabolic shift to promote lymphomagenesis. In addition to its role in metabolism, recent studies have shown that PKM2 promotes oncogenesis by phosphorylating nuclear STAT3 (signal transducer and activator of transcription 3) and regulating transcription of genes involved in cell survival and proliferation. We hypothesized that identification of novel PKM2 interactors could provide additional insights into its expanding functional role in cancer. To this end, immunocomplexes of FLAG-tagged PKM2 were isolated from NPM-ALK-positive ALCL (anaplastic large cell lymphoma) cells and subjected to liquid chromatography tandem mass spectrometry (LC–MS/MS) which led to the identification of polypyrimidine tract-binding protein (PTBP1) as a novel interactor of PKM2. The interaction between PTBP1 and PKM2 was restricted to the nucleus and was dependent on NPM-ALK mediated Y105 phosphorylation of PKM2. Stable shRNA-mediated silencing of PTBP1 resulted in a marked decrease in pY105-PKM2 and pY705-STAT3 which led to decreased ALCL cell proliferation and colony formation. Overall, our data demonstrate that PTBP1 interacts with PKM2 and promotes ALCL oncogenesis by facilitating PKM2-dependent activation of STAT3 within the nucleus.
Similar content being viewed by others
Main
A subset of anaplastic large cell lymphoma (ALCL) harbors the chromosomal translocation t(2;5)(p23;q35)1, 2 that results in the expression of nucleophosmin (NPM)-anaplastic lymphoma kinase (ALK).3 The constitutively active tyrosine kinase (NPM-ALK) promotes oncogenesis by phosphorylating proteins that regulate cell survival, invasion, and proliferation.4, 5, 6, 7, 8, 9 Currently, selective inhibition of ALK has shown to be effective for tumors that are dependent on ALK signaling including ALCL, lung adenocarcinomas, and inflammatory myofibroblastic tumors.10, 11, 12, 13, 14
NPM-ALK promotes a metabolic shift towards biomass production that favors cell proliferation(4), an emerging cancer hallmark that was initially described by Warburg et al.15 In ALCLs, this is in part a result of inactivation of the enzyme, pyruvate kinase M2 (PKM2) by tyrosine phosphorylation (Y105-PKM2).4, 16, 17 In its active tetrameric state, PKM2 converts phosphoenolpyruvate (PEP) to pyruvate in the final step of glycolysis.18 Phosphorylation of PKM2 by NPM-ALK favors the inactive dimeric isoform of PKM2 which predominates over the active tetrameric isoform,19, 20, 21 favoring biomass accumulation.16, 17
Interestingly, recent studies indicate that PKM2 may have a broader role in oncogenesis as dimeric PKM2 in the nucleus can function as a protein kinase22 and phosphorylate STAT3 (signal transducer and activator of transcription 3) which subsequently promotes gene transcription promoting enhanced cell proliferation.22, 23, 24
To elucidate novel functional roles of PKM2, we used liquid chromatography tandem mass spectrometry (LC–MS/MS) to establish a PKM2 ‘interactome’ which led to the identification of polypyrimidine tract-binding protein 1 (PTBP1). PTBP1 functions primarily as a splicing factor in the nucleus and has been implicated in cancer cell growth and tumorigenesis.25, 26 However, its expression and functional role in lymphomagenesis has not been previously described. Our studies demonstrate that the interaction of PKM2 and PTBP1 occurs predominantly in the nucleus and is dependent on NPM-ALK mediated phosphorylation of Y105-PKM2. Deregulation of the PKM2-PTBP1 complex is associated with decreased pY705-STAT3. Furthermore, knockdown of PTBP1 led to decreased pY705-STAT3 levels, cell proliferation and colony formation indicating that the interaction of PKM2-PTBP1 regulates the nuclear activation of STAT3.
Materials and methods
Cell Culture
NPM-ALK-positive ALCL-derived cell lines (DEL, SUPM2, SUDHL1) were maintained in RPMI-1640 with 10% fetal bovine serum in humidified incubators containing 5% CO2. Stable cell lines were generated by lentiviral transduction. Transduced cell lines were selected with puromycin.
Compounds
The 7-amino-1,3,4,5-tetrahydrobenzoazepinone derivative selective ALK inhibitor CEP26939 (CEP) was acquired from Cephalon.27 The ALK inhibitor Crizotinib was purchased from Sigma-Aldrich.
Antibodies and Immunoprecipitation
Protein from cell lysates were analyzed via sodium dodecyl sulfate polyacrylamide gel electrophoresis. Immunoprecipitation was performed using a FLAG-M2 peptide. Antibodies used include ALK (Invitrogen, Carlsbad, CA, USA), pY1604-ALK, PKM2, pY105-PKM2, PTBP1, STAT3, and pY705-STAT3 (Cell Signaling Technologies, Danvers, MA, USA).
Cytoplasmic/Nuclear Fractionation
ALCL cells were collected, washed, and re-suspended in 1x hypotonic buffer (20 nM Tris–HCL pH7.4, 10 mM NaCl, 2 mM MgCl2) and set on ice for 15 min. A 10% NP4O detergent was added and vortexed for 10 s before centrifugation. The resulting supernatant was used as the cytoplasmic fraction. The pellet was re-suspended in Cell Extraction Buffer (ThermoFisher Scientific, Waltham, MA, USA) and set on ice for 30 min while vortexing at 10 min intervals. The resulting solution was centrifuged and the supernatant was used as the nuclear fraction.
Protein Identification by Mass Spectrometry
Purified immunocomplexes were resolved on a SDS-PAGE gel and proteins were visualized using Colloidal Coomassie stain. Whole lane was cut into 14 pieces each and de-stained with 30% methanol for 4 h. Upon reduction (10 mM DTT) and alkylation (65 mM 2-Chloroacetamide) of the cysteines, proteins were digested overnight with sequencing grade, modified trypsin (Promega). Resulting peptides were resolved on a nano-capillary reverse phase column (Picofrit column, New Objective) using a 1% acetic acid/acetonitrile gradient at 300 nl/min and directly introduced into a linear ion-trap mass spectrometer (LTQ Orbitrap XL, ThermoFisher). Data-dependent MS/MS spectra on the 5 most intense ions from each full MS scan were collected (relative CE ~35%). Proteins were identified by searching the data against the human protein database (Uniprot, release 2011-05) appended with decoy (reverse) sequences using the X!Tandem/Trans-Proteomic Pipeline (TPP) software suite. All peptides and proteins with a PeptideProphet and ProteinProphet probability score of >0.9 (fdr <2%) were considered positive identifications and potential candidates for verification.
WST-1 and Colony Formation Assays
Cell proliferation was assessed via the WST-1 assay over a 72-h period in 3 ml of RPMI-1640 with 1% fetal bovine serum (Roche Applied Science, Indianapolis, IN, USA). Colony formation assays were performed with Methocult H4230 (Stemcell Technologies, Vancouver, British Columbia, Canada). Colonies were grown for 14 days before staining with p-iodonitrotetrazolium chloride (Sigma-Aldrich, St. Louis, MO, USA).
Immunohistochemical Studies
Tissue samples were obtained from the archives of the Hematopathology Section, Department of Pathology, University of Michigan, with institutional approval (HUM00023256). IHC staining was conducted with the following antibodies: ALK (mouse, 1:100, Dako, Carpinteria, CA, USA), PTBP1 (goat, 1:200, Abcam, Cambridge, MA, USA).
Statistical Analysis
Statistical significance was assessed using a two-tailed Student’s t-test after appropriate transformations were applied to account for normality and equality of variances. Values lower than 0.05 were considered significant. Unless otherwise specified, data are represented as mean±s.e.m.
Results
PTBP1 Interacts with PKM2 and the Interaction is Dependent on NPM-ALK Mediated Phosphorylation of Y105-PKM2
In order to gain a better understanding of PKM2’s role in ALCL oncogenesis, we used mass spectrometry to identify proteins that interact with PKM2. An ALCL cell line (DEL) was stably transduced with FLAG-tagged PKM2 (both wild-type (WT) PKM2 and non-phosphorylation mutant Y105F-PKM2) (Supplementary Figure 1A). Immunoprecipitation of PKM2 using a FLAG-M2 peptide was performed using 25 μg of protein lysate from each cell line. Non-transduced DEL cells were used as a negative control. Analysis of the mass spectrometry data revealed 63 candidate interacting proteins for WT-PKM2 and 70 for Y105F-PKM2 after subtracting those identified in the negative control (non-transduced DEL). These proteins included B-catenin and fructose 1,6-bisphosphate (FBP) (Supplementary Table 1), both known interactors of PKM2.28, 29 The candidate list also included several nuclear proteins such as HMGA1 and HNRNPU. Interestingly, most of these were identified predominantly in the WT-PKM2 immunocomplex and not in the Y105F-PKM2 immunocomplex (Supplementary Table 1).
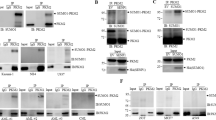
Notably, PTBP1 (Figure 1a) was identified in the WT-PKM2 immunocomplex with two unique spectral counts but not in the Y105F-PKM2 immunocomplex (Supplementary Table 1). Because of its known role in PKM1 and PKM2 splicing30 as well as its overexpression in several types of cancers,31, 32, 33 we hypothesized that its interaction with PKM2 may have a role in the pathogenesis of ALCL.
PTBP1 (polypyrimidine tract-binding protein 1) interacts with PKM2 (pyruvate kinase M2), and this interaction is dependent on NPM-ALK (nucleophosmin-anaplastic lymphoma kinase) phosphorylation at Y105-PKM2. (a) Identification of PTBP1 as a potential PKM2 interactor using LC–MS/MS (liquid chromatography tandem mass spectrometry) analysis: MS/MS spectra of 326IAIPGLAGAGNSVLLVSNLNPER348 (precursor [MH] +2=1138.49 m/z) derived from PTBP1 (SwissProt # P26599) is shown. Observed b- and y-ions are indicated. (b and c) PKM2 and its phosphorylated pY105-PKM2 isoform specifically interacts with PTBP1. Lysates from ALCL (anaplastic large cell lymphoma)-derived DEL cell lines were immunoprecipitated with anti-PKM2 (b) or anti-PTBP1 (c) antibodies before immunoblot with PTBP1, PKM2 or phosphorylation specific antibody against Y105-PKM2. Immunoprecipitation with isotype IgG controls (IP IgG) was used as negative control. The first lane represents whole-cell extract from DEL cell lines (input control). (d) PTBP1 specifically interacts with phosphorylated Y105-PKM2. Lysates from DEL cell lines with ectopic stable expression of recombinant FLAG-tagged WT-PKM2 or phosphorylation mutant Y105F-PKM2, were immunoprecipitated with anti-FLAG antibody before immunoblot with PTBP1, PKM2 or anti pY105-PKM2 antibodies. Parental DEL cell lines with no expression of FLAG-tagged vectors were used as negative controls. (e) NPM-ALK kinase activity is required for PTBP1 and PKM2 interaction. Lysates from DEL cell lines were immunoprecipitated with anti-PKM2 antibody after inhibition of NPM-ALK kinase activity with 300 nM of CEP compound or DMSO (control). The immunoprecipitates were immunblotted with antibodies against PTBP1, pY105-PKM2 and PKM2. Successful inhibition of NPM-ALK kinase activity was confirmed with specific antibodies against pY1604-ALK, the total levels of NPM-ALK were not modified during the inhibition treatment (6 h). Figures are representative pictures from at least three independent experiments.
In order to validate the interaction between PTBP1 and PKM2, immunoprecipitation of endogenous PKM2 was performed in ALCL cell lines (DEL and SUPM2) using an anti-PKM2 antibody and probed for PTBP1 by western blot analysis. PTBP1 is pulled down by PKM2 but not by the IgG control pull down (Figure 1b; Supplementary Figure 1B). Reciprocal immunoprecipitation with anti-PTBP1 antibody and western blot analysis (Figure 1c; Supplementary Figure 1C) further demonstrates that PTBP1 interacts with PKM2.
In order to determine whether phosphorylation of Y105-PKM2 is necessary for its interaction with PTBP1, FLAG-tagged WT-PKM2 and Y105F-PKM2 were overexpressed in DEL cells and lysates were subjected to immunoprecipitation with a FLAG-M2 peptide. Immunoprecipitation with IgG and non-transduced DEL cells acted as negative controls. As shown in Figure 1d, PTBP1 is pulled down by WT-PKM2 but not by Y105F-PKM2. These data are consistent with the results of our mass spectrometry experiments (Supplementary Table 1) and suggests that the phosphorylation of PKM2 at Y105 is important for its interaction with PTBP1.
As phosphorylation of PKM2 at Y105 is regulated by NPM-ALK,4 we hypothesized that NPM-ALK activity regulates the interaction of PKM2 with PTBP1. To investigate this, lysates from DEL cells subjected to either a small molecule inhibitor of ALK (CEP) or DMSO (control) for 6 h were immunoprecipitated with PKM2 and analyzed by western blot. As shown in Figure 1e and Supplementary Figure 1D, inhibition of NPM-ALK activity in both cell lines (DEL and SUDHL1) led to significant ablation of the interaction of PTBP1 with PKM2. This observation supports the hypothesis that NPM-ALK-dependent phosphorylation of PKM2 at Y105 regulates its interaction with PTBP1.
PTBP1 Interacts with pY105-PKM2 in the Nucleus
To further understand the functional significance of our observations, we assessed the subcellular location of the interaction between PTBP1 and PKM2. Cytoplasmic and nuclear subfractions of ALCL cell lines (DEL, SUPM2, and SUDHL1) were enriched and HSP70 and LaminB2 western blots were used to assess the efficiency of cytoplasmic and nuclear enrichment, respectively. PKM2 is localized predominantly in the cytoplasm with low levels found in the nucleus (Figure 2a). In contrast, PTBP1 localization is abundant within the nuclear subfraction with lower amounts in the cytoplasm (Figure 2a). The isolated subcellular fractions were then subjected to immunoprecipitation with anti-PKM2-antibody and analyzed by western blot analysis (Figure 2a; Supplementary Figure 1E). Interestingly, the interaction of PTBP1 with PKM2 was detected in the nuclear fraction and not in the cytoplasmic fraction (Figure 2a; Supplementary Figure 1E). To confirm this finding, reciprocal immunoprecipitation using an anti-PTBP1-antibody was performed (Figure 2b; Supplementary Figure 1F). Analysis of the PTBP1 immunocomplex corroborated the previous observations and showed that the interaction of PKM2 and PTBP1 occurs predominantly in the nuclear fraction (Figure 2b). Interestingly, pY105-PKM2 was not detected in the immunoprecipitation of the cytoplasmic fraction suggesting that PTBP1 preferentially interacts with pY105-PKM2 within the nucleus.
PTPB1 (polypyrimidine tract-binding protein 1) interacts with PKM2 (pyruvate kinase M2) preferentially in the nucleus over the cytoplasm. (a and b) PTBP1 and PKM2 predominantly interact in nuclear fractions of ALCL (anaplastic large cell lymphoma)-derived cell lines. Nuclear and cytoplasmic fractions of ALCL-derived DEL cell lines were immunoprecipitated with anti-PKM2 (a) or anti-PTBP1 (b) antibodies, before immunoblot with PKM2, phosphorylation specific antibody against Y105-PKM2 and PTBP1 antibodies. Successful nuclear/cytoplasmic fractionation was confirmed by selective expression of HSP70 and LaminB2 in cytoplasmic and nuclear fractions, respectively. Immunoprecipitation with isotype IgG antibody was used as non-specific binding control. (c) Interaction between phosphorylated Y105-PKM2 and PTBP1 predominantly occurs in nuclear fractions of ALCL-derived cell lines. Nuclear and cytoplasmic fractions from lysates of DEL cell lines with ectopic stable expression of recombinant FLAG-tagged WT-PKM2 or phosphorylation mutant Y105F-PKM2, were immunoprecipitated with anti-FLAG antibody before immunoblot with PTBP1, PKM2 or anti pY105-PKM2 antibodies. Figures are representative pictures from at least three independent experiments.
To provide additional evidence that PTBP1 interacts with pY105-PKM2 preferentially in the nucleus, the interaction was evaluated in subcellular fractions of DEL cell lines stably expressing either FLAG-tagged WT-PKM2 or Y105F-PKM2. Consistent with previous observations, the interaction of PTBP1 and pY105-PKM2 was only detected in the nuclear subfraction (Figure 2c). Furthermore, the interaction was abolished in cells expressing Y105F-PKM2. Overall, these data indicate that PTBP1 interacts with pY105-PKM2 in the nucleus, and suggests that the PTBP1-PKM2 complex may be relevant to a nuclear function of PKM2.
PTBP1 Regulates PKM2-Dependent Phosphorylation of Y705-STAT3 in the Nucleus
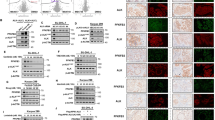
PKM2 has been reported to exhibit protein kinase activity against STAT3 in the nucleus in several types of cancers.22 To determine whether the interaction of PTBP1 with Y105-PKM2 affects its ability to regulate STAT3 activation, we assessed the levels of Y705-STAT3 in ALCL-derived DEL cells in the presence or absence of CEP, a selective inhibitor of ALK as well as in DEL cells stably expressing either WT-PKM2 or Y105F-PKM2. As seen in Figure 3a, pY1604-ALK and pY105-PKM2 levels decreased upon ALK inhibition (CEP treatment). Analysis of the subcellular fractions showed that nuclear levels of phosphorylated PKM2 were completely abolished upon NPM-ALK inhibition. Interestingly, expression of pY705-STAT3 in the nucleus was abolished in cells expressing Y105F-PKM2 (Figure 3b). This provides evidence that Y105-PKM2 has an important role in regulating the phosphorylation of Y705-STAT3.
PTBP1 (polypyrimidine tract-binding protein 1) regulates the PKM2 (pyruvate kinase M2)-dependent phosphorylation of Y705-STAT3 (signal transducer and activator of transcription 3) in the nucleus. (a) Nuclear pY105-PKM2 and pY705-STAT3 decrease upon selective inhibition of NPM-ALK (nucleophosmin-anaplastic lymphoma kinase) kinase activity. Lysates from nuclear and cytoplasmic fractions of DEL cell lines with selective NPM-ALK inhibition (CEP) or control (DMSO) were immunblotted with antibodies against PTBP1, pY105-PKM2, PKM2, STAT3, and pY705-STAT3. Successful inhibition of NPM-ALK kinase activity was confirmed with specific antibodies against pY1604-ALK, the total levels of NPM-ALK, STAT3, PTBP1, and PKM2 were not modified during the inhibition treatment (300 nM CEP for 6 h). Successful nuclear/cytoplasmic fractionation was confirmed by selective expression of HSP70 and LaminB2 in cytoplasmic and nuclear fractions, respectively. (b) Nuclear pY705-STAT3 decreases upon inhibition of Y105-PKM2 phosphorylation. Nuclear and cytoplasmic fractions from lysates of DEL cell lines with ectopic stable expression of recombinant FLAG-tagged WT-PKM2 or phosphorylation mutant Y105F-PKM2, were immunoblotted with antibodies against PTBP1, pY105-PKM2, PKM2, STAT3, and pY705-STAT3. Total levels of STAT3, PTBP1, and PKM2 were not modified upon ectopic expression of WT-PKM2 or Y105F-PKM2. (c) Inhibition of nuclear NPM-ALK kinase activity decreases the nuclear localization of pY105-PKM2 and pY705-STAT3. The nuclear fractions from DEL cell lines were treated with the specific NPM-ALK kinase inhibitor Crizotinib (300 nM - Criz) or DMSO for 6 h, before immunoblot with antibodies against PTBP1, pY105-PKM2, PKM2, STAT3, and pY705-STAT3. (d) PTBP1 regulates the phosphorylation pY105-PKM2 and pY705-STAT3. Lysates from DEL cell lines with stable expression of PTBP1 shRNA (PTBP1 KD) or scramble shRNA were immunoblotted with antibodies against PTBP1, pY105-PKM2, PKM2, PKM1, STAT3, pY705-STAT3, NPM-ALK, and pY1604-ALK. Numbers indicate integrated signal density of overlying western blot signal bands, stable shRNA against PTBP1 decreased its protein expression by ~80% in comparison with scramble controls. Cell lines with stable expression of PTBP1 shRNA show decreased expression of pY705-STAT3 (~50%) and pY105-PKM2 (~60%). No significant modification in the expression of WT-PKM2, WT-PKM1, WT-STAT3, NPM-ALK, or pY1604-ALK was identified. Anti-ACTIN was used as loading control. (e) PTBP1 regulates the nuclear localization of pY105-PKM2 and pY705-STAT3. Nuclear and cytoplasmic lysates from DEL cell lines with stable expression of PTBP1 shRNA (PTBP1 KD) or scramble shRNA were immunoblotted with antibodies against PTBP1, pY105-PKM2, PKM2, STAT3, and pY705-STAT3. Total levels of WT-PKM2 and WT-STAT3 were not modified in the nuclear or cytoplasmic fractions upon selective knockdown or PTBP1. Figures are representative pictures from experiments performed more than 3 times.
The localization of NPM-ALK within the nucleus has been previously described,34 however its role in regulating the phosphorylation of PKM2 and STAT3 within the nucleus has not been addressed. In order to determine whether NPM-ALK phosphorylates Y105-PKM2 within the nucleus, nuclear subfractions of DEL cell lysates were treated with an ALK inhibitor (Crizotinib) which exhibits similar level of ALK inhibition as CEP26939 (CEP).4, 35 As shown in Figure 3c, inhibition of NPM-ALK kinase resulted in decreased levels of total PKM2, pY105-PKM2, and pY705-STAT3. Total levels of NPM-ALK and STAT3 remained unchanged.
To determine whether PTBP1 regulates the kinase activity of PKM2, we assessed Y705-STAT3 in DEL cells after stable expression of shRNA against PTBP1 using scramble shRNA as a negative control. As shown in Figure 3d, stable knockdown of PTBP1 (79% of control) did not affect the expression of endogenous PKM2 and PKM1 (Figure 3d). However, a significant reduction of pY105-PKM2 (57%) and pY705-STAT3 (46%) was observed after PTBP1 knockdown suggesting that PTBP1 may regulate the levels of Y105-PKM2. These data suggest that PTBP1 regulates PKM2-dependent phosphorylation of Y705-STAT3. Notably, this is independent of ALK activity as the levels of Y1604-ALK were not affected.
To determine whether PTBP1 affects PKM2-dependent phosphorylation of STAT3, we analyzed subcellular fractions of DEL cells after shRNA-mediated knockdown of PTBP1. In contrast to the control (scramble shRNA) in which levels of pY705-STAT3 were higher in the nucleus than in the cytoplasm, cells transduced with PTBP1 shRNA, showed complete abolishment of phosphorylated Y705-STAT3 specifically in the nucleus (Figure 3e). Total STAT3 levels were relatively equivalent in both fractions (Figure 3e). These observations support the hypothesis that PTBP1 modulates the levels of Y105-PKM2 and Y705-STAT3 within the nucleus.
PTBP1 Promotes cell Growth and Proliferation
STAT3 is an important oncogenic mediator of ALCL.6 Our results indicate that pY105-PKM2 and PTBP1 both modulate the levels of Y705-STAT3 in the nucleus. Thus, we hypothesized that PTBP1 may have a role in oncogenesis of ALCL. Evaluation of cell proliferation and colony formation capacity of DEL cells with stable knockdown of PTBP1 (Figure 3d) showed significant decrease in the number of colonies relative to the scramble expressing control cells 14 days post-seeding (Figure 4a). Cellular proliferation was also significantly decreased after 24 h in cells in which PTBP1 was knocked down in comparison to the scramble control (Figure 4b). These data provide evidence that PTBP1 has a significant role in promoting oncogenesis. To further support the functional role of PTBP1 in ALCLs, we carried out immunohistochemical studies for PTBP1 in formalin fixed paraffin-embedded tissue biopsies of ALK-positive ALCLs. As shown in Supplementary Figure 2, we observed strong nuclear PTBP1 in 8/8 cases (100%) of ALK-positive ALCLs providing support for a functional role in these neoplasms.
PTBP1 (polypyrimidine tract-binding protein 1) promotes cell growth and proliferation. (a) PTBP1 regulates anchorage-independent growth of ALCL (anaplastic large cell lymphoma)-derived cell lines. ALCL-derived DEL cell lines with stable expression of PTBP1 shRNA or scramble shRNA were allowed to grow in methycult media for 14 days, and subsequently were stained with iodonitrotetrazolium chloride (INT dye). The number of colonies in each group was counted; bar graphs represent mean±standard error mean (SEM) from at least three independent experiments. T-test statistical analysis was performed with *P-value<0.01. (b) PTBP1 regulates the proliferation of ALCL-derived cell lines. Proliferation rate of ALCL-derived DEL cell lines with stable expression of PTBP1 shRNA or scramble shRNA was measured in 24 h intervals for a total of 72 h. Line graph represent mean±s.e.m. of WST-1 reagent absorbance (viability) at each time point, from at least three independent experiments. T-test statistical analysis was performed with *P-value<0.05.
Discussion
Recent studies have shown that the enzymatically inactive isoform of PKM2 has an oncogenic role in the cell nucleus, activating downstream targets involved in survival and proliferation. Phosphorylation of PKM2 at Y105 by NPM-ALK defines a dual oncogenic role for PKM2; generating biomass in the cytoplasm and upregulating gene transcription in the cell nucleus. Prior studies22 have demonstrated that the homodimeric isoform of PKM2 predominantly localizes to the cell nucleus. Nuclear expression of PKM2 is also observed in ALCL cell lines, however further biochemical studies are required to determine whether it exists in a homodimeric conformation in ALCL cells. Importantly, this is the first study to show that expression and interaction of PTBP1 with PKM2 in the nucleus has an oncogenic role in lymphomas.
Analysis of the proteomic and biochemical data identified PTBP1 as a key partner for PKM2 in the nucleus. The interaction of PTBP1 and PKM2 in the nucleus regulates the phosphorylation and activation of STAT3, which is a key mediator of oncogenesis in ALCL. As this interaction is restricted to the nucleus, proper translocation of PKM2 from cytoplasm to the nucleus is a key step for the oncogenic role of PKM2. Moreover, our data indicate that the nuclear kinase activity of NPM-ALK regulates the pY105-PKM2 in the nucleus and thus PKM2-dependent phosphorylation of STAT3.
The molecular mechanism by which PTBP1 potentiates PKM2 kinase activity in the nucleus stills remains to be elucidated. Previous reports have shown that PTBP1 promotes transcription of PKM2 over the less oncogenic splice isoform PKM1.25 Downregulation of PTBP1 could prevent the transcription of PKM2 and therefore decrease its kinase activity in the nucleus. Another possible explanation to consider would be that the PKM2-PTBP1 protein complex may enhance the kinase activity of PKM2 secondary to conformational changes. Future studies to elucidate the role of the interaction between PTBP1 and PKM2 are warranted.
Our findings show that in ALCL cells, PTBP1 is critical for the activation of STAT3 in the nucleus (Figure 5), which is highly active in many cancers and an emerging therapeutic target.36 PTBP1 has also been found to be upregulated in several types of cancers, and our findings suggest that it may be used as a potential therapeutic target.25 Further explorations into the potential therapeutic advantages of targeting PTBP1 are warranted.
Proposed mechanism for PTBP1 (polypyrimidine tract-binding protein 1) mediating PKM2 (pyruvate kinase M2)-dependent activation of STAT3 (signal transducer and activator of transcription 3) in the nucleus. Phosphorylation of pY105-PKM2 by NPM-ALK (nucleophosmin-anaplastic lymphoma kinase) modifies PKM2’s conformational status from a tetrameric to a dimeric state, which facilitates its nuclear localization shift. Within the nucleus, dimeric PKM2 interacts with PTBP1 to mediate the phosphorylation of Y705-STAT3 and promoting ALCL (anaplastic large cell lymphoma) anchorage-independent growth and proliferation. In addition, nuclear kinase activity of NPM-ALK regulates protein levels of PKM2 and ultimately increase available pool of phosphorylated Y105-PKM2 at the nucleus.
References
Morris SW, Kirstein MN, Valentine MB et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science 1994;263:1281–1284.
Falini B . Anaplastic large cell lymphoma: pathological, molecular and clinical features. Br J Haematol 2001;114:741–760.
Stein H, Mason DY, Gerdes J et al. The expression of the Hodgkin's disease associated antigen Ki-1 in reactive and neoplastic lymphoid tissue: evidence that Reed-Sternberg cells and histiocytic malignancies are derived from activated lymphoid cells. Blood 1985;66:848–858.
McDonnell SR, Hwang SR, Rolland D et al. Integrated phosphoproteomic and metabolomic profiling reveals NPM-ALK-mediated phosphorylation of PKM2 and metabolic reprogramming in anaplastic large cell lymphoma. Blood 2013;122:958–968.
McDonnell SR, Hwang SR, Basrur V et al. NPM-ALK signals through glycogen synthase kinase 3beta to promote oncogenesis. Oncogene 2012;31:3733–3740.
Zamo A, Chiarle R, Piva R et al. Anaplastic lymphoma kinase (ALK) activates Stat3 and protects hematopoietic cells from cell death. Oncogene 2002;21:1038–1047.
Zhang Q, Raghunath PN, Xue L et al. Multilevel dysregulation of STAT3 activation in anaplastic lymphoma kinase-positive T/null-cell lymphoma. J Immunol 2002;168:466–474.
Amin HM, McDonnell TJ, Ma Y et al. Selective inhibition of STAT3 induces apoptosis and G(1) cell cycle arrest in ALK-positive anaplastic large cell lymphoma. Oncogene 2004;23:5426–5434.
Chiarle R, Simmons WJ, Cai H et al. Stat3 is required for ALK-mediated lymphomagenesis and provides a possible therapeutic target. Nat Med 2005;11:623–629.
Gambacorti-Passerini C, Messa C, Pogliani EM . Crizotinib in anaplastic large-cell lymphoma. N Engl J Med 2011;364:775–776.
Carpenter EL, Mosse YP . Targeting ALK in neuroblastoma—preclinical and clinical advancements. Nat Rev Clin Oncol 2012;9:391–399.
Murga-Zamalloa C, Lim MS . ALK-driven tumors and targeted therapy: focus on crizotinib. Pharmgenomics Pers Med 2014;7:87–94.
Butrynski JE, D'Adamo DR, Hornick JL et al. Crizotinib in ALK-rearranged inflammatory myofibroblastic tumor. N Engl J Med 2010;363:1727–1733.
Kwak EL, Bang YJ, Camidge DR et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363:1693–1703.
Warburg O, Wind F, Negelein E . The metabolism of tumors in the body. J Gen Physiol 1927;8:519–530.
Christofk HR, Vander Heiden MG, Wu N et al. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature 2008;452:181–186.
Hitosugi T, Kang S, Vander Heiden MG et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal 2009;2:ra73.
van Berkel TJ, de Jonge HR, Koster JF et al. Kinetic evidence for the presence of two forms of M2-type pyruvate kinase in rat small intestine. Biochem Biophys Res Commun 1974;60:398–405.
Schneider J, Neu K, Grimm H et al. Tumor M2-pyruvate kinase in lung cancer patients: immunohistochemical detection and disease monitoring. Anticancer Res 2002;22:311–318.
Eigenbrodt E, Reinacher M, Scheefers-Borchel U et al. Double role for pyruvate kinase type M2 in the expansion of phosphometabolite pools found in tumor cells. Crit Rev Oncog 1992;3:91–115.
Zwerschke W, Mazurek S, Massimi P et al. Modulation of type M2 pyruvate kinase activity by the human papillomavirus type 16 E7 oncoprotein. Proc Natl Acad Sci USA 1999;96:1291–1296.
Gao X, Wang H, Yang JJ et al. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell 2012;45:598–609.
Gao X, Wang H, Yang JJ et al. Reciprocal regulation of protein kinase and pyruvate kinase activities of pyruvate kinase M2 by growth signals. J Biol Chem 2013;288:15971–15979.
Li Q, Zhang D, Chen X et al. Nuclear PKM2 contributes to gefitinib resistance via upregulation of STAT3 activation in colorectal cancer. Sci Rep 2015;5:16082.
He X, Arslan AD, Ho TT et al. Involvement of polypyrimidine tract-binding protein (PTBP1) in maintaining breast cancer cell growth and malignant properties. Oncogenesis 2014;3:e84.
Izaguirre DI, Zhu W, Hai T et al. PTBP1-dependent regulation of USP5 alternative RNA splicing plays a role in glioblastoma tumorigenesis. Mol Carcinog 2012;51:895–906.
Ott GR, Tripathy R, Cheng M et al. Discovery of a potent inhibitor of anaplastic lymphoma kinase with in vivo antitumor activity. ACS Med Chem Lett 2010;1:493–498.
Yang W, Xia Y, Ji H et al. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature 2011;480:118–122.
Jurica MS, Mesecar A, Heath PJ et al. The allosteric regulation of pyruvate kinase by fructose-1,6-bisphosphate. Structure 1998;6:195–210.
Calabretta S, Bielli P, Passacantilli I et al. Modulation of PKM alternative splicing by PTBP1 promotes gemcitabine resistance in pancreatic cancer cells. Oncogene 2016;35:2031–2039.
Sugiyama T, Taniguchi K, Matsuhashi N et al. MiR-133b inhibits growth of human gastric cancer cells by silencing pyruvate kinase muscle-splicer polypyrimidine tract-binding protein 1. Cancer Sci 2016;107:1767–1775.
Guo J, Jia J, Jia R . PTBP1 and PTBP2 impaired autoregulation of SRSF3 in cancer cells. Sci Rep 2015;5:14548.
Taniguchi K, Sugito N, Kumazaki M et al. MicroRNA-124 inhibits cancer cell growth through PTB1/PKM1/PKM2 feedback cascade in colorectal cancer. Cancer Lett 2015;363:17–27.
Greenland C, Touriol C, Chevillard G et al. Expression of the oncogenic NPM-ALK chimeric protein in human lymphoid T-cells inhibits drug-induced, but not Fas-induced apoptosis. Oncogene 2001;20:7386–7397.
Murga-Zamalloa CA, Mendoza-Reinoso V, Sahasrabuddhe AA et al. NPM-ALK phosphorylates WASp Y102 and contributes to oncogenesis of anaplastic large cell lymphoma. Oncogene, advance online publication, 3 October 2016.
Chai EZ, Shanmugam MK, Arfuso F et al. Targeting transcription factor STAT3 for cancer prevention and therapy. Pharmacol Ther 2015;162:86–97.
Acknowledgements
This work was supported by the Department of Pathology, University of Michigan and R01 CA140806-01 (MSL), R01 DE119249, R01 CA136905 (KSJ-EJ).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Laboratory Investigation website
This study describes the interaction between pyruvate kinase M2 (a critical regulator of glycolysis) and PTBP1 (polypyrimidine tract-binding protein), and outlines the characteristics of this interaction as well as the role the proteins play in promoting downstream signaling and oncogenesis in NPM-ALK-positive anaplastic large cell lymphoma (ALCL). This evidence suggests that PTBP1 is a potential therapeutic target in ALCL.
Rights and permissions
About this article
Cite this article
Hwang, S., Murga-Zamalloa, C., Brown, N. et al. Pyrimidine tract-binding protein 1 mediates pyruvate kinase M2-dependent phosphorylation of signal transducer and activator of transcription 3 and oncogenesis in anaplastic large cell lymphoma. Lab Invest 97, 962–970 (2017). https://doi.org/10.1038/labinvest.2017.39
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2017.39
This article is cited by
-
PTBP1 drives c-Myc-dependent gastric cancer progression and stemness
British Journal of Cancer (2023)
-
Pyruvate kinase M1 regulates butyrate metabolism in cancerous colonocytes
Scientific Reports (2022)
-
18F-FDG PET/CT imaging findings in anaplastic large cell lymphoma, a rare subtype of lymphoma
Cancer Imaging (2020)
-
Protein kinase function of pyruvate kinase M2 and cancer
Cancer Cell International (2020)
-
Existence of reprogrammed lymphoma stem cells in a murine ALCL-like model
Leukemia (2020)