Abstract
Colorectal cancer arises in part from the cumulative effects of multiple gene lesions. Recent studies in selected cancer types have revealed significant intra-tumor genetic heterogeneity and highlighted its potential role in disease progression and resistance to therapy. We hypothesized the existence of significant intra-tumor genetic heterogeneity in rectal cancers involving variations in localized somatic mutations and copy number abnormalities. Two or three spatially disparate regions from each of six rectal tumors were dissected and subjected to the next-generation whole-exome DNA sequencing, Oncoscan SNP arrays, and targeted confirmatory sequencing and analysis. The resulting data were integrated to define subclones using SciClone. Mutant-allele tumor heterogeneity (MATH) scores, mutant allele frequency correlation, and mutation percent concordance were calculated, and copy number analysis including measurement of correlation between samples was performed. Somatic mutations profiles in individual cancers were similar to prior studies, with some variants found in previously reported significantly mutated genes and many patient-specific mutations in each tumor. Significant intra-tumor heterogeneity was identified in the spatially disparate regions of individual cancers. All tumors had some heterogeneity but the degree of heterogeneity was quite variable in the samples studied. We found that 67–97% of exonic somatic mutations were shared among all regions of an individual’s tumor. The SciClone computational method identified 2–8 shared and unshared subclones in the spatially disparate areas in each tumor. MATH scores ranged from 7 to 41. Allele frequency correlation scores ranged from R2=0.69–0.96. Measurements of correlation between samples for copy number changes varied from R2=0.74–0.93. All tumors had some heterogeneity, but the degree was highly variable in the samples studied. The occurrence of significant intra-tumor heterogeneity may allow selected tumors to have a genetic reservoir to draw from in their evolutionary response to therapy and other challenges.
Similar content being viewed by others
Main
Colorectal cancer is the third leading cause of cancer-related death in the US, and rectal cancers comprise about a third of the burden of colorectal cancer.1 Treatment of rectal cancer requires complex multimodal therapy because of the increased risk of local recurrence as compared with colon cancer.2 Response to the pre-operative combination chemotherapy and radiation therapy is variable with up to 30% of patients demonstrating a complete pathological response.3, 4 No particular clonal somatic mutations or biomarkers have been found that predict these differences in response; however, the studies to date have not been properly powered or comprehensive.5 We hypothesized that rectal cancers may exhibit significant intra-tumor genetic heterogeneity and that this heterogeneity may have relevance in therapeutic response and/or tumor recurrence.
Intra-tumor genetic heterogeneity, such as manifested by heterogeneity in point mutations or copy number changes among cancer cells, has been described in several tumor types, including lung cancer,6, 7 renal cell carcinoma,8 chronic lymphocytic leukemia,9 breast cancer,10, 11 and acute myeloid leukemia.12 The heterogeneity reflects the presence of different subclonal populations within the cancer and likely impacts the patient’s clinical course and response to therapy. Defining subclonal populations within solid tumors is challenging and requires costly and complex analysis and interpretation methods. Hence, some groups have utilized other mathematical approaches to assess and describe heterogeneity.13, 14 With the exception of a very recent publication revealing substantial intra-tumor heterogeneity in localized mutations and copy number changes between individual glands in adenomas and colon cancers,15 prior publications describing intra-tumor heterogeneity in colorectal cancer have reported the existence of heterogeneity, but the depth of the analyses was limited to studies at a few genetic loci.16, 17
The nature of intra-tumor heterogeneity can be defined and described in multiple different ways, including considerations of differences in localized mutations (point or small insertion/deletion mutations), and differences in copy number changes, as well as by a calculation known as the mutant allele tumor heterogeneity, MATH score.14, 18 In patients with head and neck squamous cell carcinomas, high MATH scores were associated with poor outcome, whereas low MATH scores are associated with better outcomes. Whether these measures of heterogeneity correlate with each other or with the extent of subclonality in a tumor is unknown.
We present here an in-depth assessment of intra-tumor heterogeneity in six rectal cancers. We performed the next-generation Illumina whole-exome sequencing of normal rectal mucosa and two or three spatially distinct regions of rectal cancers from six patients, obtaining median gene coverage per sample of 47-fold. We then used the Ion Torrent platform to perform targeted re-sequencing for validation allowing us to obtain an average depth of greater than 400-fold per gene. We in addition utilized Oncoscan SNP arrays to assess for copy number and allele frequency changes. We examined heterogeneity with regard to single-nucleotide variants, insertions, deletions, allele frequency correlations, copy number changes and correlations, MATH scores, and percent mutation correlation. We combined the deep, targeted sequencing and the copy number changes data using a previously described computational method called SciClone to estimate the number of subclones within the tumors.14 We found rectal cancers exhibit intra-tumor genetic heterogeneity as well as evidence of subclones as assessed by multiple measures and the degree of heterogeneity varied considerably among patients.
MATERIALS AND METHODS
Sample Collection and DNA Extraction
For five of the rectal cancers, DNA was isolated from banked surgical resection materials present in the University of Michigan Cancer Center Tissue Procurement Core. A qualified gastrointestinal pathologist assessed multiple independent regions from each of the five primary rectal cancers. Two or three spatially distinct areas of adenocarcinoma were identified in each cancer specimen via inspection of hematoxylin- and eosin- (H&E)-stained glass slides. Each tumor region intended for analysis was at least 1 cm in distance from the other area(s). Normal rectal tissue was also collected from the surgical specimens. The relevent area of each frozen tumor blocks were identified and the normal and tumor material were manually dissected from the blocks. For one patient’s specimens, spatially distinct areas of tumor and normal rectum were removed from the in situ tumor at the time of ultrasound and the tissues were frozen. These samples were also assessed for tumor purity by a gastrointestinal pathologist via analysis of representative H&E-stained slides. Clinical data on each patient was abstracted from the medical records. The University of Michigan Institutional Review Board approved this study. DNA was extracted from frozen tissue, using the Allprep mini kit (Qiagen, Valencia, CA, USA) and according to the manufacturer’s instructions.
Next-Generation Sequencing
Illumina whole-exome sequencing and analysis
Genomic DNA samples were fragmented using a Covaris S2 fragmentation system to a target size of 300 bp. The samples were end-repaired, a-tailed, and custom adapters were ligated using the NEBNext DNA Library Prep kit according to the manufacturers recommended protocols. The custom adapters included 6 bp barcodes designed using BARCRAWL software19 and synthesized by Integrated DNA Technologies. After ligation, the samples were size selected to 300 bp on a 2% agarose gel and 1- mm gel slices were retained. Samples were isolated from the gel using the Qiagen QIAquick gel extraction system. Either 10 or 15 μl of each ligation product was enriched using the Phusion master mix kit and custom PCR primers for a total of 14 cycles of PCR amplification. The PCR products were purified using AmpureXP beads.
Library QC was performed using the Agilent Bioanalyzer and qPCR. Each pool was captured using the Nimblegen SeqCap EZ V3 Exome Enrichment Kit according to the manufacturer’s recommended protocols. The capture pools were combined and sequenced on the Illumina HiSeq 2000 platform with paired-end 100 bp reads using v3 reagents.
Paired reads were mapped to the hg19 reference genome using the Burrows–Wheeler Aligner20 and were processed with SAMtools.21 Duplicates were removed with the rmdup function and variants were called with the mpileup function. Variants were identified as on-target if they fell within 10 bp of a targeted area defined by the exome-capture panel, and off-target variants removed. The samples were processed by mpileup as normal-tumor pairs so that the likelihood ratio of each variant could be estimated.22 Variants identified only in the tumor sample with a likelihood ratio (CLR value) >60 were considered somatic, representing <1 in 1 million likelihood of a false somatic identification. Somatic variants identified in one region of the tumor but not the others were considered unique to that region. Variants were then annotated with SnpEff23 to classify variants by region (e.g., exon, intron, 5′-UTR), to determine amino acid change type (e.g., synonymous and non-synonymous) and to prioritize according to the predicted impact of the variant. Variants within non-coding transcripts (e.g., pseudogenes), and variants present in dbSNP v.135, were removed. Depth of coverage was calculated as number of reads aligned to exons in a gene multiplied by the length of reads per length of all exons in gene. We then calculated the median coverage per gene per sample.
Ion Torrent Sequencing and Analysis
Variants identified from Illumina sequencing data were targeted for sequencing by Ion Torrent using custom Ion Torrent Ampliseq panels. Briefly, for each targeted variant, a single-overlapping amplicon (~100 bp) was designed, and assigned to one of two separate targeted panels for sequencing. Ampliseq library preparation with barcode incorporation, template preparation, and sequencing using the Ion Torrent PGM was performed as described.24 Custom VCF files containing all targeted variants were used for variant calling using command-line Torrent Variant Caller (TVC, v4.2.3) with default somatic low-stringency settings after alignment by TMAP v3.6.3 in Torrent Suite v3.6.2. VCF-level filtering of candidate variant calls generated by TVC was applied to remove variant calls with flow-corrected read depth (FDP) <20, flow-corrected variant allele containing reads (FAO) <6, variant fraction of <5%, variants in homopolymer runs≥4, or those with extreme skewing of forward/reverse flow-corrected reads supporting the variant allele (FSAF/FSAR <0.2 or >5). Base-level filtering was then applied to candidate somatic variants passing aforementioned criteria to exclude additional technical artifacts, including removal of variants located at the last mapped base (or outside) of amplicon target regions and variants with the majority of supporting reads harboring excess additional mismatches or indels (likely sequencing error). Variants called in 30% or more of samples sequenced on a given targeted panel were removed as panel-specific technical artifacts. Variants present at allele frequencies >0.5% in ESP6500 or 1000 Genomes Project (reported by wAnnovar25) were filtered out as known polymorphisms. To restrict our focus exclusively to somatic variation, we also filtered out variants present in any of the matched or unmatched normal samples. We have previously shown similar filtering criteria identifies variants passing Sanger sequencing validation with >95% accuracy.26, 27, 28 A subset of called variants was inspected by visualizing the alignment bam files and corresponding variant.vcf files in the GenomeBrowse genome browser software (Golden Helix). APC indels were assessed manually.
Oncoscan SNP Arrays and Copy Number Variation Analysis
Affymetrix OncoScan V3 arrays were run on all tumor samples. The OncoScan array platform consists of a set of 217 k probes designed specifically for profiling tumors. The assay detects copy number change by generating data at 50–100 kb resolution across a set of 891 cancer genes, and 300–400 kb across the rest of the genome. Raw array florescence intensity data generated on the Affymetrix scanners in the form of CEL files were loaded into the OncoScan Console software v.1.1.0 (Affymetrix, Santa Clara, CA, USA). Quality control statistics as well as integrated OSCHP files were generated by OncoScan Console. The standard Affymetrix reference control file for OncoScan data was used for processing the arrays.
The SNP-FASST2 algorithm implemented in the Nexus software v.7.5 (BioDiscovery, El Segundo, CA, USA) was used to make copy number calls. SNP-FASST2, rather than reporting on the large number of individual probe locations across the genome, uses a Hidden Markov Model based approach to identify larger copy number segments based on a log-ratio threshold derived from all probes in a given region. Ratios are the log2 ratios of the normalized intensity of the sample over the normalized intensity of a reference with further correction for a sample-specific variation. The Median Log2 Ratio is computed for each segment detected in the analysis. The significance threshold for segmentation was set at 1.0e−5 also requiring a minimum of three probes per segment and a maximum probe spacing of 1000 kbp between adjacent probes before breaking a segment. Segments were classified as having gains when the Log2Ratio (L2R) exceeded 0.2, losses when L2R was <−0.2, and with high copy gains and homozygous losses being called when L2R was >0.6 and <−1.0, respectively. Differences in copy number changes between samples from a tumor for individual genes were counted when one sample had a change as defined above and another either did not have that change or had a different change.
B-allele frequency information was also generated for each tumor. Median BAF is reported for each segment and is the median BAF of the markers identified as heterozygous. If the number of heterozygous markers in the segment is below 10 or the percent of homozygous markers is above 85% no value is reported. The B-allele frequency values are used to determine whether a segment is in a loss of heterozygosity (LOH) or an allelic imbalance state. By default, probe sets were automatically centered to the median for all samples by the Nexus software. For individual samples where the median probe set value was not diploid, specified regions of balanced heterozygosity were manually identified by visual inspection of L2R and BAF plots and defined as diploid regions, permitting the Nexus software to reset the entire probe set to the newly defined areas.
We calculated Copy Number Correlations by comparing copy number changes between pairs of samples within each tumor. We correlated the median centered (tumor/normal) L2R values over the chromosomal regions (Table 3 and Supplement 3). We computed copy number estimates for individual genes as the median copy number estimate over the segments spanned by the gene’s transcript. We called copy number estimates as different in two samples if the log2 estimates differed by at least 0.2 units.
MATH and Correlations
In order to calculate the MATH score, the distributions of alternate (mutant) allele frequencies for all somatic variants detected in each sample were plotted using the R plot and density functions. The MATH score, a robust version of the coefficient of variation, was computed for each sample as 100 times the median absolute deviation from the median (the MAD) divided by the median: MATH=100* MAD/median.14, 18 Tumor purity is corrected for in this calculation as it is both in the numerator and the denominator.
We calculated the mutation percent concordance between every pair of samples within a tumor by computing 100%*S/(S+((U1+U2)/2)), where S is the number of shared mutations observed in both of the two samples and U1 and U2 are the number of mutations found in one sample but not the other, for each of the two samples, respectively.13 The presence of other samples for the tumor has no effect on the calculation. We also calculated allele frequency correlations by computing the (Pearson) correlation between mutant allele frequencies (obtained via Ion Torrent) for every pair of samples from the same tumor, over the 24 910 genes interrogated by exome sequencing.
SciClone Analysis
SciClone is an R package developed to infer subclonal populations of cells in a tumor sample.14 The software implements a variational Bayesian mixture model to classify variants into different populations based on their copy number states and allele frequencies, with clusters in the diploid copy number state being potential subclones. A probability is calculated for the presence of each variant in each of the inferred subclones. The validated variant allele frequencies and read depths from the Ion Torrent analysis of each tumor and the copy number and LOH data from the OncoScan arrays were used as input into SciClone, generating cluster models for the tumors.
Data Sharing
The Ion Torrent sequencing data can be found at the European Variation Archive website: http://www.ebi.ac.uk/eva/?eva-study=PRJEB10956. The OncoScan array data are available from the NCBI’s Gene Expression Omnibus29 at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE73365 accession number GSE73365.
RESULTS
Patient Characteristics
The clinical features of the six patients whose cancers were analyzed are summarized in Table 1. Five of the six tumors were collected as surgical resection specimens in patients who did not undergo pre-operative chemotherapy and radiation. Samples from NP1 were prospectively collected at the time of his endorectal ultrasound before radiation and chemotherapy. However, the pathological features described represent those in the surgical resection of his primary tumor following 6 months of combination 5-fluorouracil, leucovorin, and oxaliplatin and then combination therapy with xeloda and 50.4 Gy of radiation over 6 weeks. Each of the spatially distinct regions subjected to analysis in each patient’s tumor were assessed by a gastrointestinal pathologist and showed similar histological features.
Variants Identified
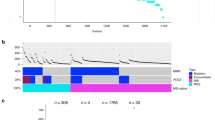
Utilizing Illumina whole-exome sequencing, we identified 333 somatic, coding variants across our samples. An average of the median gene coverage per sample of 47-fold was obtained by Illumina sequencing across exonic regions, with the lowest depth of 19-fold (patient 204, Ca2) and the highest of 89-fold (patient NP1, Ca2). Ion Torrent sequencing of the entire targeted panel was performed on each sample, achieving an average read depth of 405-fold per target across all samples, with the lowest average depth being 165-fold (patient 20, Ca1) and the highest 698-fold (patient 20, normal). We identified and validated between 30 and 89 somatic variants per tumor (Table 2). This is similar to the number of variants identified per tumor in The Cancer Genome Atlas (TCGA) studies of colon and rectal cancers, where the mean number of somatic mutations in non-hypermutated tumors was 58.30 From one to three of the genes identified as being somatically mutated in tumor samples of each of the six patients were previously noted to be significantly mutated in the TCGA study of colorectal cancers (Table 2 and Figure 1).30
Allele frequencies of coding variants in each tumor and sample in six rectal cancers. Mutations that failed validation with Ion Torrent were not included. Red indicates genes listed as significant in TCGA colorectal cancer paper. Yellow indicates high impact mutations: frameshift, splice site, and stop codons. Blue indicates synonymous mutations. Black indicates the mutation was not found in that sample. S, sample; TCGA, The Cancer Genome Atlas.
Intra-Tumor Heterogeneity in Variants
An initial assessment, using only the Illumina exome-capture data, of presence and absence of specific somatic variants suggested that a given tumor shared as little as 23% of the variants among all samples (data not shown). Targeted, higher depth re-sequencing did not validate this degree of heterogeneity in all samples but did still reveal substantial mutational heterogeneity. Overall, the false negative rate for variants identified using Illumina sequencing and then tested using Ion Torrent was 38% and the false positive rate was 8%. The false negative Illumina calls were identified when the Illumina sequencing identified a variant in one sample from a tumor but failed to call a variant in another sample from the same tumor. Had we failed to assess the variants with deeper sequencing analysis, we would have dramatically overestimated the degree of intra-tumor heterogeneity in the samples. However, because low-depth Illumina sequencing was used to identify the variants for targeted sequencing, we are likely under estimating heterogeneity.
When Ion Torrent-validated mutations were assessed, the percentage of all coding validated mutations that were shared among all of the spatially distinct regions of an individual tumor ranged between 67 and 97% (Table 2 and Figure 1). Mutation percent concordance was calculated for every pair of distinct samples within each tumor and ranged from 86.5 to 98.7% (Supplement 1 and Table 3). Allele frequency correlations were calculated for the spatially distinct samples within each tumor revealing R2=0.75–0.94 (Supplement 1 and Table 3).
MATH
Another means of describing intra-tumor genetic heterogeneity is by generating a histogram or density plot of the mutant allele frequencies and calculating a score based on the ratio of the width of the distribution to the center of the distribution.14 Heterogeneous tumors will tend to exhibit a broader allele frequency distribution due to genomically distinct cellular populations, centered at a lower allele fraction, than a homogeneous tumor (Figure 2). MATH scores and density plots were calculated for each sample from each tumor (Figure 2, Table 2 and, figure Supplement 2). The resulting MATH values ranged from 7 to 42. The density plots indicate somewhat narrow, unimodal distributions of allele frequencies in Tumor 4 and Tumor 11 (Figure 2, Supplement 1). Peaks immediately flanking the modal peak can theoretically be explained by single chromosomal duplication (three-copy genotype regions), with smaller peaks lacking a matching equivalent on the opposite side of the modal peak, or asymmetrical shoulders to the modal peak, representing possible subclonal contributions. A peak present at double the allele frequency of the main peak may be caused by regions of single-copy deletion (one-copy genotype regions). The median allele frequency distribution peak for these tumors is between 0.3 and 0.4, with the exception of 11-Ca3, which has a median ~0.2. An ideal distribution of purely heterozygous mutations would be expected to center ~0.5; however, contaminating normal DNA or non-aberrant cells present in the tumor have the effect of shifting the median to a lower value. Tumors 1 and 20 exhibit more broad distribution profiles with more marked multimodality (Figure 2, Supplement 2). The two tumor profiles of patient 204 display radically different density profiles, indicating a possible sampling of different subclonal populations or potential significant differences in tumor purity between samples (Supplement 2).
Intra-Tumor Heterogeneity in Copy Number
Differences in copy number between samples for the same tumors can be reflected in heterogeneity measures like the MATH scores, since variant frequencies are affected, but direct copy number change measurements provide additional orthogonal information about genomic alteration events. It is well known that multiple genomic regions can differ from the diploid state in colorectal cancers.30, 31 Copy number changes were assessed using OncoScan Arrays (Figure 3a and Supplement 3). Several quality control metrics are generated by the OncoScan Console software for assessing the performance of each array assay, the two main metrics being MAPD and ndSNPQC which measure noisiness of the log2ratio and the differentiation of the B-allele frequency tracks, respectively. Arrays with MAPD ≤0.3 and ndSNPQC ≥26 are considered ‘within bounds’. Out of the 15 arrays generated in this experiment, 10 arrays were within bounds and 5 were outside bounds (4Ca1, 204Ca1, NP1Ca1-3). Profiles for the outside bounds arrays were determined to be useful for overall visual comparison with the caveat that specific calls and boundaries of copy number regions for these arrays are less precisely determined. The overall reported percentage of aberrant cells varied from 40 to 80 percent, with the percentage undeterminable for 7 arrays. Manual interpretation of the undetermined arrays based on the log2R and B-allele frequency plots indicate aberrant cell percentages between 30 and 57 percent. Several tumor samples were indicated by the OncoScan Console software (using the TuScan algorithm) as being almost entirely non-diploid, with overall ploidy of 4 (Tumor 1, Tumor 20Ca2) (Figure 3,Supplement 3). OncoScan Console was unable to determine correct ‘normal diploid’ regions for these tumors and for one other tumor of undetermined ploidy (204Ca1). For these samples, normal diploid regions were identified manually within the Nexus Copy Number software, permitting recalculation of profiles for these tumors using the SNP-FASST2 algorithm.
Intra-tumor Heterogeneity in copy number changes. (a) Heterogeneity in copy number changes in three samples from Tumor 20. (b) Tumor 20 shows heterogeneity in chromosome 3, in which regions of the p and q arms vary between states of allelic balance and imbalance across the three samples, as evidenced by characteristic three band or four band patterns in the BAF plots.
Significant regions of copy number change were detected in all tumors, with several tumors exhibiting quite complex copy number profiles (Tumors 20 and 204). All six tumors displayed significant gains of chr20q, with all but Tumor 4 having gains of variable length of chr8q. Losses were observed on chr18 in all samples, except in Tumor 204, and for chr8p in Tumor 11, Tumor 20, and NP1. Tumor 20 shows heterogeneity in chromosome 3, in which regions of the p and q arms vary between states of allelic balance and imbalance across the three samples, as evidenced by characteristic three band or four band patterns in the BAF plots, respectively (Figure 3b). Care must be taken in interpreting these plots so as not to confuse differences in tumor purity with copy number change or allelic differences; most differences in the Tumor 20 profiles may be attributed to estimated aberrant cells fractions of 70, 90, and 50% for 20Ca1, 20Ca2, and 20Ca3, respectively (Figure 3a). Examples of intra-tumor heterogeneity were observed in some tumors such as Tumor 11 where differences for gains of chr8q were seen in Ca2 vs Ca1 and Ca3 as well as in losses of 8p, where the first ~24 Mb are commonly deleted but the rest of the deleted segment varies between the samples (Supplement 3). Similarly, for Tumor 1, a large portion of chr3 is lost in Ca1 but not in Ca2 (Supplement 3). The copy number changes found in individual samples from the same tumor were variably correlated, showing correlations from 0.74 to 0.93 (Table 3 and Supplement 1). Regions of copy number change were assessed specifically at the sites of the 32 genes highlighted as being significant in TCGA publication characterizing colorectal cancer.30 Depending on the tumor assessed between 0 and 66% of the 32 TCGA specified genes were found to have differences in copy number changes between pairs of samples for the same tumor. The average was 19.5% with binomial 95% confidence interval of 15.6–23.5%. This was compared with differences in copy number between the samples for 100 random genes where we found differences in 17.0% of the pairs of samples (95% confidence interval: 2.0–62.0%). Differences between pairs for the 32 highlighted genes was highly correlated to the number of differences found in the 100 random genes across pairs of samples (r=0.95, P=1 × 10−6). We found that the frequency of copy number changes in the TCGA genes is no different than other genes (P=0.26, Z-test of two proportions).
SciClone
The high-depth targeted sequencing results and copy number variations were used to identify subclones in the diploid portions of the tumors using the previously published method called SciClone.14 The R package utilizes the copy number changes to identify mutations that are only in the diploid region of the tumor and then the algorithm defines subclones by clustering variant allele frequencies. The SciClone plots display subclones within each tumor, some of which are shared between the spatially distinct samples while others are not (Figure 4 and Supplement 4). For Tumor 1 (Figure 4a), SciClone identified three distinct subclones in the copy number 2 portion of the tumor. These were shared equally between samples from the tumor. However, in Tumor 11, (Figure 4b), SciClone identified six distinct subclones in the copy number 2 portion of the tumor. The number and distribution of subclones varied in the six tumors studied (Supplement 4).
SciClone plots: (a) Tumor 1, VAF is plotted against density and the copy number 2 variants are identified along the green line. Then VAF is plotted against tumor coverage (depth) for the two samples from Tumor 1 for variants found in the diploid portion of the samples and subclones are defined by different colors. VAFs for each sample are then plotted against each other in the diploid portion to reveal the relationship between the multiple clusters (subclones). (b) Tumor 11; Tumor 11, VAF is plotted against density and the copy number 2 variants are identified along the green line. Then VAF is plotted against tumor coverage (depth) for the three samples from Tumor 11 for variants found in the diploid portion of the samples and subclones are defined by different colors. VAFs for each sample are then plotted against each other in the diploid portion to reveal the relationship between the multiple clusters (subclones). VAF, variant allele frequency.
Differences in Heterogeneity
Some of the tumors studied manifested high levels of intra-tumor genetic heterogeneity in the spatially distinct samples from the tumors, using each of the analysis strategies. Other tumors manifested considerably less intra-tumor genetic heterogeneity. For example, Tumor 4 had 93% of its mutations shared across the spatially distinct regions of the tumor tested (Figure 1 and Table 2), as well as a dominant peak on its allele frequency density plot with near complete overlap between the two spatially disparate areas (Figure 2). In addition, tumor 4 had low MATH scores (7 and 9), high allele frequency correlation (0.910), mutation percent concordance (96.3 percent), and moderate change in copy number with copy number correlation between samples of 0.802 and only three subclones identified by SciClone (Tables 2 and 3, Supplement 1 and 4). Tumors demonstrating higher amounts of heterogeneity are Tumor 20 and Tumor 11. Tumor 11 shares only 67% of mutations between all spatially disparate samples (Table 1) and the correlation between mutant allele frequencies was 0.750. MATH scores for Tumor 20 were very high, perhaps due to a greater number of high frequency mutant allele fractions, which increases the MATH score even if observed in every sample (Table 2, Supplement 2). There were six subclones identified on the SciClone analysis in Tumors 11 and Tumor 8 in Tumor 20. Most correlations between measures of heterogeneity are not significant likely because of our small sample size (Table 3, Supplement 1). However, the allele frequency correlation is correlated to the concordance, as expected (r=0.89, P=0.017).
DISCUSSION
The studies and data presented here offer strong evidence of intra-tumor genetic heterogeneity in rectal cancer. The heterogeneity observed among spatially distinct regions in a given tumor was found for localized mutations, allele frequency measures, MATH scores, as well as copy number alterations. In addition, the computational method, SciClone, identified multiple subclones in the tumors, some of which were shared among the different samples from a given tumor and the others were private. Genetic heterogeneity was confirmed using high-depth DNA sequencing coverage provided by Ion Torrent-based analyses. Our study utilizes more in-depth sequencing analyses for the six rectal cancers studied than some of the prior work examining colorectal cancer intratumoral genetic heterogeneity. As in other types of cancers, intra-tumor heterogeneity is likely to be of potential biological and clinical significance in rectal cancer.
Our findings are largely in agreement with those in a recent study of individual glands in adenomas and colon cancers that revealed substantial intra-tumor heterogeneity in the targeted panel of mutations and copy number alterations leading the authors to present a ‘Big Bang’ model whereby there are early expansions of a large number of subclones that intermix.15 Sottoriva et al sequenced both bulk colon tumors and individual tumor glands and found evidence of early and widespread intra-tumor heterogeneity. We exclusively used rectal cancers in our studies. Colon and rectal cancers are treated differently and multiple studies have suggested they are different genetic entities, thus it is important to study each of them in order to understand how their genetics may predict their phenotype and response to therapy. Other prior studies to address intra-tumor heterogeneity in colorectal cancer have been more limited. Losi et al assessed between 9 and 14 areas of 45 colorectal cancers for specific somatic mutations in TP53 and KRAS along with LOH of chromosomes 5q and 18q. They found evidence of intra-tumor heterogeneity for at least one marker tested in 76% of tumors.17 Diaz et al found evidence for increasing levels of mutant KRAS alleles in the circulation of CRC patients whose primary tumors were largely comprised of wild-type KRAS alleles, when the patients were treated with anti-EGFR therapy. The authors performed mathematical modeling suggesting that cells with these KRAS mutant alleles were present at low levels in the tumors before treatment and then expanded in the setting of biological selection during anti-EGFR therapy.32 The findings highlight the potentially significant role for intra-tumor heterogeneity in the response or resistant to current and future therapies.33 Historically, APC, KRAS, and TP53 have been thought of as key driver mutations in colorectal cancer development.34 Over time, with further sequencing, other drivers have been identified.30, 35 As expected, we found some of these potential alternative driver mutations in our samples. As explained by Lawrence, Getz, and colleagues in their recent manuscript on driver mutations, there are likely many more driver gene lesions to be found.36
Our data regarding shared and unique mutations and copy number alterations among spatially distinct regions of individual tumors implies that distinct subclones are frequently present in rectal tumors. Limitations of this study include: low depth of our initial sequencing and use of normal adjacent rectum as our control. The initial Illumina sequencing results that formed the basis for the targeted panel were relatively low coverage (average of 47-fold). Greater initial sequencing depth may have identified more low allele frequency variants, some of which may be early events private to a particular location or help to define additional subclones. However, the depth used to define our panel is similar to that used in other studies. For example, Sottoriva et al used 20–60 × coverage Illumina whole-exome sequencing to define the targets of their Ion Torrent panel.15 In addition, we used DNA from normal appearing nearby rectum as our normal control rather than DNA from blood. It is possible that DNA from this tissue may not perfectly represent normal due to passenger mutations in the mucosa that have not resulted in a neoplastic phenotype, but these likely are present at a very-low allele frequency because any given mutation is not likely to be in the entire area samples due to the long time to division for any given crypt.37
Despite its apparent clinical importance in head and neck cancer, the MATH score approach to assess intra-tumor genetic heterogeneity in rectal cancer may have limitations. If in all samples for a hypothetical cancer, half the mutant genes have a mutant allele fraction of 0.45 and half have 0.90 (due to loss of the wild-type alleles), the MATH score is 33.3, which would be rather large. However, in this same hypothetical cancer, the concordance and allele frequency correlations would both be 1.0, and indeed we would not say we could see any heterogeneity in the samples. Nevertheless, in our studies, the MATH score in the tumors that were more heterogeneous by other measures, were high, and the MATH score determination is a relatively inexpensive method for assessing heterogeneity as it only requires low pass exome sequencing as compared with SciClone which requires deep sequencing and SNP arrays or equivalent copy number data for copy number changes, so it may have utility. In addition, if a tumor is a mixture of many different clones but they are spatially well mixed, different samples may yield nearly identical variant allele frequencies and copy number data, yet the MATH score will detect the mixture.
We calculated four different measures of intra-tumor heterogeneity and identified subclones using SciClone. As of yet, we do not know which will be clinically important or whether they will correlate with each other. These measures vary between the different tumors. More research is needed to develop clinically useful metrics from mutation frequency and heterogeneity data.
The potential clinical significance of intra-tumor genetic heterogeneity for cancer prognosis or for treatment of colorectal cancer patients has yet to be explored. Studies in head and neck cancer have shown that increased MATH scores are a poor prognostic indicator.18 In chronic lymphocytic leukemia, Landau et al found that patients treated with chemotherapy underwent expansion of resistant subclones and that the presence of subclonal driver mutations was an independent predictor of rapid disease progression.9 These studies suggest that intra-tumor heterogeneity may be a genetic reservoir to be drawn from in times of stress and that the extent and nature of intra-tumor genetic heterogeneity should be examined further for its clinical significance in patients with colon and rectal cancer.
References
Siegel R, Naishadham D, Jemal A . Cancer statistics, 2012. CA Cancer J Clin 2012;62:10–29.
van Gijn W, Marijnen CA, Nagtegaal ID et al. Preoperative radiotherapy combined with total mesorectal excision for resectable rectal cancer: 12-year follow-up of the multicentre, randomised controlled TME trial. Lancet Oncol 2011;12:575–582.
Habr-Gama A, de Souza PM, Ribeiro U Jr et al. Low rectal cancer: impact of radiation and chemotherapy on surgical treatment. Dis Colon Rectum 1998;41:1087–1096.
Garcia-Aguilar J, Hernandez de Anda E, Sirivongs P et al. A pathologic complete response to preoperative chemoradiation is associated with lower local recurrence and improved survival in rectal cancer patients treated by mesorectal excision. Dis Colon Rectum 2003;46:298–304.
Garcia-Aguilar J, Chen Z, Smith DD et al. Identification of a biomarker profile associated with resistance to neoadjuvant chemoradiation therapy in rectal cancer. Ann Surg 2011;254:486–492.
Zhang J, Fujimoto J, Wedge DC et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science 2014;346:256–259.
de Bruin EC, McGranahan N, Mitter R et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 2014;346:251–256.
Gerlinger M, Rowan AJ, Horswell S et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883–892.
Landau DA, Carter SL, Stojanov P et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 2013;152:714–726.
Nik-Zainal S, Alexandrov LB, Wedge DC et al. Mutational processes molding the genomes of 21 breast cancers. Cell 2012;149:979–993.
Nik-Zainal S, Van Loo P, Wedge DC et al. The life history of 21 breast cancers. Cell 2012;149:994–1007.
Ding L, Ley TJ, Larson DE et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 2012;481:506–510.
Torres L, Ribeiro FR, Pandis N et al. Intratumor genomic heterogeneity in breast cancer with clonal divergence between primary carcinomas and lymph node metastases. Breast Cancer Res Treat 2007;102:143–155.
Miller CA, White BS, Dees ND et al. SciClone: inferring clonal architecture and tracking the spatial and temporal patterns of tumor evolution. PLoS Comput Biol 2014;10:e1003665.
Sottoriva A, Kang H, Ma Z et al. A Big Bang model of human colorectal tumor growth. Nat Genet 2015;47:209–216.
Baldus SE, Schaefer KL, Engers R et al. Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clin Cancer Res 2010;16:790–799.
Losi L, Baisse B, Bouzourene H et al. Evolution of intratumoral genetic heterogeneity during colorectal cancer progression. Carcinogenesis 2005;26:916–922.
Billich A . Thymosin alpha1. SciClone Pharmaceuticals. Curr Opin Investig Drugs 2002;3:698–707.
Frank DN . BARCRAWL and BARTAB: software tools for the design and implementation of barcoded primers for highly multiplexed DNA sequencing. BMC Bioinformatics 2009;10:362.
Li H, Durbin R . Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–1760.
Li H, Handsaker B, Wysoker A et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078–2079.
Li H . A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011;27:2987–2993.
Cingolani P, Platts A, Wang le L et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92.
McDaniel AS, Zhai Y, Cho KR et al. HRAS mutations are frequent in inverted urothelial neoplasms. Hum Pathol 2014;45:1957–1965.
Chang X, Wang K . wANNOVAR: annotating genetic variants for personal genomes via the web. J Med Genet 2012;49:433–436.
Cani AK, Hovelson DH, McDaniel AS et al. Next-Gen Sequencing Exposes Frequent MED12 Mutations and Actionable Therapeutic Targets in Phyllodes Tumors. Molecular Cancer Res 2015;13:613–619.
Hovelson DH, McDaniel AS, Cani AK et al. Development and validation of a scalable next-generation sequencing system for assessing relevant somatic variants in solid tumors. Neoplasia 2015;17:385–399.
Warrick JI, Hovelson DH, Amin A et al. Tumor evolution and progression in multifocal and paired non-invasive/invasive urothelial carcinoma. Virchows Arch 2015;466:297–311.
Edgar R, Domrachev M, Lash AE . Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207–210.
Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330–337.
Xie T, D’Ario G, Lamb JR et al. A comprehensive characterization of genome-wide copy number aberrations in colorectal cancer reveals novel oncogenes and patterns of alterations. PLoS One 2012;7:e42001.
Diaz LA Jr., Williams RT, Wu J et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 2012;486:537–540.
Crockford A, Jamal-Hanjani M, Hicks J et al. Implications of intratumour heterogeneity for treatment stratification. J Pathol 2014;232:264–273.
Commo F, Ferte C, Soria JC et al. Impact of centralization on aCGH-based genomic profiles for precision medicine in oncology. Ann Oncol 2015;26:582–588.
Cosentino F, Arena L, Banchini L et al. Epidemiologic surveillance in occupational bladder cancer: a Tuscan experience. G Ital Med Lav Ergon. 2007 Jul -Sep; 29:313–315.
Lawrence MS, Stojanov P, Mermel CH et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014;505:495–501.
Baker AM, Cereser B, Melton S et al. Quantification of crypt and stem cell evolution in the normal and neoplastic human colon. Cell Rep 2014;8:940–947.
Acknowledgements
Translational Research in GI Cancer: 5P50CA130810, American Surgical Association Foundation Fellowship; Cancer Center Core Grant: 5P30A046592.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Laboratory Investigation website
In this study of 6 rectal cancers, the authors show that spatially distinct regions of individual tumors share as few as 67% of their mutations and that there are multiples differences in copy number changes across regions. The SciClone algorithm identified multiple shared and unshared genetic subclones within each tumor. The degree of intra-tumor heterogeneity varies substantially between tumors and may play a role in disease progression and resistance to therapy.
Supplementary information
Rights and permissions
About this article
Cite this article
Hardiman, K., Ulintz, P., Kuick, R. et al. Intra-tumor genetic heterogeneity in rectal cancer. Lab Invest 96, 4–15 (2016). https://doi.org/10.1038/labinvest.2015.131
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2015.131
This article is cited by
-
MRI radiomics independent of clinical baseline characteristics and neoadjuvant treatment modalities predicts response to neoadjuvant therapy in rectal cancer
British Journal of Cancer (2022)
-
Overexpression of the cohesin-core subunit SMC1A contributes to colorectal cancer development
Journal of Experimental & Clinical Cancer Research (2019)
-
Cytolytic activity correlates with the mutational burden and deregulated expression of immune checkpoints in colorectal cancer
Journal of Experimental & Clinical Cancer Research (2019)
-
A microsatellite based multiplex PCR method for the detection of chromosomal instability in gastric cancer
Scientific Reports (2018)
-
Utilization of the oncoscan microarray assay in cancer diagnostics
Applied Cancer Research (2017)