Abstract
The specific mechanisms that mediate CD4+ T-cell-mediated liver injury have not been fully elucidated. CD4+ invariant natural killer T (iNKT) cells are required for liver damage in some mouse models of hepatitis, while the chemokine receptors CXCR3 and CCR5 are considered dominant Th1 chemokine receptors involved in Th1 trafficking in inflammatory conditions. BALB/c-Tgfb1−/− mice spontaneously develop Th1 hepatitis. Here, we directly test the hypotheses that iNKT cells or the Th1-cell chemokine receptors CXCR3 and CCR5 are required for development of liver disease in Tgfb1−/− mice. Tgfb1−/− mouse livers exhibited significant increases in iNKT cells and in ligands for CXCR3 or CCR5. Tgfb1−/− mice were rendered deficient in iNKT cells, CXCR3, CCR5, or both CXCR3 and CCR5, by cross-breeding with appropriate knockout mice. Tgfb1−/− mice developed severe liver injury, even in the absence of functional CD1d/iNKT cells, CXCR3, CCR5, or both CXCR3 and CCR5. Liver CD4+ T cells accumulated to high numbers, and spleen CD4+ T-cell numbers declined, regardless of the functionality of the CXCR3/CCR5 response pathways. Similarly, dendritic cells and macrophages accumulated in Tgfb1−/− livers even when CXCR3 and CCR5 were knocked out. Th1-associated cytokines (IFN-γ, TNF-α, IL-2) and chemokines (CXCL9, CXCL10) were strongly overexpressed in Tgfb1−/− mice despite knockouts in CD1d, CXCR3, or CCR5. These studies indicate that the cellular and biochemical basis for CD4+ T-cell-mediated injury in liver can be complex, with myriad pathways potentially involved.
Similar content being viewed by others
Main
Autoimmune hepatitis (AIH) and viral hepatitis are inflammatory liver diseases in which liver damage is dependent to a great extent upon the infiltration of liver by CD4+ T cells producing the cytokine IFN-γ,1, 2, 3 also known as Th1 cells. In these hepatitides, Th1 CD4+ T-cell production of IFN-γ strongly correlates with disease progression and liver injury.2, 4 The use of laboratory mice has advanced the understanding of the pathogenesis of inflammatory liver diseases, as findings in laboratory mice can delineate possible mechanisms by which T cells may participate.
Liver CD4+ T cells include both conventional effector CD4+ T helper cells, and a class of non-conventional CD4+ T cells known as invariant natural killer T (iNKT) cells. iNKT cells are of particular interest in hepatitis as they are abundant in murine liver, representing between 5 and 40% of T cells found in this organ, depending upon the mouse strain.5 iNKT cells are characterized by expression of a specific TCRα chain (Vα14-Jα18) paired with a limited set of TCRβ chains. iNKT cells recognize and are activated by glycolipids presented by the MHCI-like molecule, CD1d. Upon stimulation, iNKT cells rapidly produce copious amounts of cytokines, including IFN-γ.6, 7
The involvement of iNKT cells in inflammatory liver disease was first clearly demonstrated in an experimental model of T-cell-mediated liver damage, in which Concanavalin A (ConA) is administered to mice to induce hepatitis. ConA is a T-cell lectin with predilection for the liver, in which it binds efficiently to liver sinusoidal endothelial cells.8 Administration of ConA rapidly results in T-cell activation in the liver, destruction of endothelium and the development of acute liver injury.9, 10 It was demonstrated over a decade ago that iNKT cells are among the first subset of cells to respond to ConA, resulting in rapid release of TNF-α and expression of the hepatotoxic molecule FasL.11, 12 Mice rendered unable to generate iNKT cells fail to develop liver damage following ConA administration,11, 12 demonstrating that iNKT cells are necessary for liver damage in this system. Additional studies show that acute activation of iNKT cells, via delivery of iNKT ligand α-galactosylceramide, rapidly results in cytokine release and acute liver injury in mice.13 Thus, iNKT cell activation can be both necessary and sufficient to induce liver damage in mice. The ConA model is by far the most commonly used model of CD4+ T-cell-mediated hepatitis in research, due in large part to the relative technical accessibility of this model to most research laboratories. It is critically important, however, that observations reported in the ConA model be rigorously tested in other model systems of CD4+ T-cell-mediated liver damage. Such approaches help distinguish inflammatory pathways common to immune mediated liver injury from those specific to particular inciting insults or events.
T cells migrate into sites of inflammation through the activity of chemokines. CXCR3 and CCR5 are the two principal chemokine response pathways that mediate Th1 migration in inflamed tissue, and evidence suggests that chemokines that bind to these two receptors can mediate lymphocyte migration into inflamed liver.14 In AIH patients, serum levels of IP-10 (a ligand for CXCR3) positively correlate with AST levels and with several other disease parameters,15, 16 and expression levels of CCL4 and CCL5 (which bind CCR5) are upregulated in AIH.17 Liver infiltrating lymphocytes isolated from HCV-infected livers express high levels of CCR5 and CXCR3;18 chemokines binding to CCR5 are primarily restricted to the portal tract, and CXCR3 chemokines are upregulated in the sinusoidal endothelium.18 Consistent with a role for CXCR3 in inflammatory liver disease, CXCR3 ligand expression is enhanced in HCV compared with normal liver.19 Whether one or both of these chemokine receptor pathways is critical for influx of T helper cells into liver in all types of T-cell-mediated hepatitis needs to be rigorously tested. This is particularly important to evaluate as interest in chemokine receptor antagonism as a therapeutic intervention in liver inflammation is gaining traction.20
Transforming growth factor-beta (TGF-β) is a cytokine with potent and pleiotropic effects on the immune response. TGF-β powerfully inhibits T-cell proliferation and cytokine production, antagonizes T helper cell differentiation to the Th1 and Th2 effector states, and promotes the development of regulatory T cells. TGF-β inhibits the inflammatory activities of macrophages and neutrophils, and exerts powerful regulatory effects on B cells and dendritic cells (reviewed in Li et al21). BALB/c-background mice homozygous for a null mutation in the gene encoding TGF-β1 (BALB/c-Tgfb1−/− mice) develop an aggressive Th1 CD4+ T-cell-mediated necroinflammatory liver damage, succumbing within ∼2 weeks post-natally,22 and serve as a tractable model of spontaneously developing Th1-mediated hepatocellular damage. We have previously demonstrated the pathological importance of Th1 CD4+ T cells to liver damage in this model system, as mice rendered deficient either in CD4+ T cells or IFN-γ exhibit little to no detectable liver damage.23, 24 Plasma IFN-γ levels are pathologically high in BALB/c-Tgfb1−/− mice and correlate with the extent of liver damage.24 Moreover, CD4+ T cells produce copious amounts of IFN-γ and are the only source of detectable IFN-γ production in these mice.24 There is no detectable pathological role for either CD8+ T cells or for the death receptor protein fas,24 and the production of IL-17 is undetectable (JGC and JDG, unpublished data). In this study, we further the understanding of the pathogenesis of liver damage in BALB/c-Tgfb1−/− mice by evaluating whether liver damage and inflammation are dependent upon either iNKT cells, or on CXCR3 or CCR5, the two principal chemokine receptor pathways that target Th1 cells to sites of inflammation.
MATERIALS AND METHODS
Mice
All mice were maintained at The Geisel School of Medicine at Dartmouth according to Association for Assessment and Accreditation of Laboratory Animal Care practices. BALB/c-Tgfb1−/− mice were generated as previously described.22 Cxcr3−/− mice on the BALBc background were previously described.25, 26 Cd1d−/− mice on the BALB/c-background and Ccr5−/− mice on the C57Bl/6 background were purchased from Jackson Laboratory (Bar Harbor, ME). C57Bl/6-Ccr5−/− mice were backcrossed onto the BALB/c-background in five generations using a marker assisted selection (ie, ‘speed congenic’) approach. Mouse genomes were assessed at the DartMouse Speed Congenic Core Facility at The Geisel School of Medicine at Dartmouth. DartMouse uses the Illumina (San Diego, CA) GoldenGate Genotyping Assay to interrogate 1449 SNPs spread throughout the genome. Genetic background was determined to be >99% BALB/c after the final back-cross generation. Double-knockout and triple-knockout mice were generated by cross-breeding and PCR screening for the particular knockout alleles.
Cell Isolation and Counts
Mononuclear cells were obtained from livers as previously described.27 Briefly, livers were perfused with sterile PBS, disaggregated between frosted microscope slides and filtered twice through 70 μm filters. Pellets were subjected to a 35% Percoll gradient, then RBC lysis, to obtain a single-cell suspension. Splenocytes were isolated through mechanical disruption. Cell enumeration was by trypan blue exclusion using a hemacytometer.
FACS
Single-cell suspensions were Fc blocked for 15 min at 4 °C followed by washing in PBS. Cells were then stained with anti-CD4, anti-CD3, anti-CD11b, anti-CD11c, anti-F4/80 (all from eBioscience) and/or CD1d tetramer loaded with PBS057 (NIH Tetramer Facility) or appropriate isotype control antibody or unloaded tetramer for 30 min at 4 °C, then washed and analyzed by FACS. Stained cells were analyzed on either an Accuri C6 Cytometer or BD FacsCalibur and data analyzed by CFlow or FlowJo analysis software.
Liver Lysate and Plasma Analysis by Luminex and ELISA
Protein was extracted from livers by high-speed homogenization of perfused livers in lysate buffer (0.25 M sucrose, 10 mM Tris, pH 7.4, protease inhibitors) at 120 mg (wet weight)/ml. Lysates were spun at 10 000 RCF, and supernatants collected and stored at −80 °C. Clarified supernatants, or, in some experiments, plasma, were used in Multiplex Luminex analyses performed by the Immune Monitoring Laboratory at The Geisel School of Medicine at Dartmouth or in ELISA (R&D Systems), by the manufacturer’s established protocol.
Histology and AST
Livers were perfused and fixed in PBS-buffered formalin, and prepared for H&E staining as described.22 Serum AST enzyme activity was measured as described.22
qRT-PCR
Following cardiac perfusion, total livers were homogenized in TRIzol, and RNA extracted and purified. Real-time PCR (qRT-PCR) was carried out as previously described with gene specific primers, and amplicons detected using SYBR green.28
Statistics
Statistical significance was determined by the Student’s t or Mann–Whitney tests, utilizing GraphPad Prism4 software. A P-value ⩽0.05 was considered to be significant. A P-value between 0.05 and 0.10 was considered to be approaching statistical significance.
RESULTS
T-Cell-Mediated Liver Damage in Tgfb1−/− Mice is Extensive Even in the Absence of CD1d-Restricted iNKT Cells
We previously showed that CD4+ T-cell depletion abrogates liver damage in BALB/c-Tgfb1−/− mice.23, 24 We used an anti-CD4 monoclonal antibody to deplete CD4+ T cells. This strategy targets not only conventional CD4+ T cells but also iNKT cells, which likewise express CD4. Therefore, whether iNKT cells contribute to liver damage in BALB/c-Tgfb1−/− mice is currently unknown. Mice homozygous for a null allele in the Cd1d gene do not develop iNKT cells and are a useful experimental tool to assess the requirement for iNKT cells.29
CD1d tetramer analysis readily identified iNKT cells in livers from BALB/c-Tgfb1−/− mice (Figure 1a). At post-natal day 11, when liver injury reaches peak severity,22 iNKT cells were present at higher frequency in Tgfb1−/− liver than in Tgfb1+/− littermate control liver (Figure 1b). To specifically test the requirement for iNKT cells, we bred CD1d-deficient mice with Tgfb1−/− mice to obtain mice deficient in both TGF-β1 and CD1d/iNKT cells, and then assessed liver damage at post-natal day 11. Despite an absence of CD1d/iNKT, double-knockout mice developed quite marked liver damage, measured either quantitatively by AST (Figure 1c), or qualitatively by histology (Figures 2a–c). Indeed, AST was actually somewhat higher in BALB/c-Cd1d−/−Tgfb1−/− mice (Figure 1c), suggesting that iNKT cells may have a suppressor function in this model system. Regardless, these results indicate that, distinct from the ConA model, CD1d-restricted invariant iNKT cells are not required for the development of liver damage in Tgfb1−/− mice.
iNKT cell numbers are increased in liver in Tgfb1−/− mice but do not contribute to tissue damage. (a) Liver mononuclear cells (MNC) from 11-day-old mice were stained with anti-CD3 and CD1d-αgalcer tetramer and analyzed by flow cytometry. Representative flow plots are displayed. (b) Combined data showing the percentage of CD3+ cells that stain positive for the CD1d-α-galcer tetramer. n=3 or more mice per genotype. *P<0.05, Student’s t-test. Error bars indicate s.d. (c) AST was measured from 11-day-old mice of the indicated genotypes. White asterisks reflect statistical differences (*P<0.02) between Tgfb1−/− mice and their littermate control Tgfb1+/+ mice and Tgfb1+/− mice (aggregated as ‘Tgfb1+’). n=6 mice per genotype, except for the Cd1d−/−Tgfb1+ genotype, for which n=4. NS indicates not significant. Mann–Whitney test. Error bars indicate s.d.
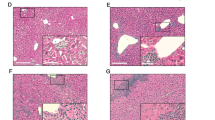
Histological liver damage in Tgfb1−/− mice is independent of CD1d/iNKT cells, CXCR3, and CCR5. Low power micrographs of H&E-stained livers from mice of the following genotypes: (a) Tgfb1+; (b) Tgfb1−/−; (c) Cd1d−/−Tgfb1−/−; (d) Cxcr3−/−Tgfb1−/−; (e) Ccr5−/−Tgfb1−/−; (f) Cxcr3−/−Ccr5−/−Tgfb1−/−. Obvious necrosis of the liver is observed in Tgfb1−/− mice, independent of the presence or absence of CD1d/iNKT cells, CXCR3, CCR5, or CXCR3/CCR5.
Massive CD4+ T-cell Accumulation and Liver Damage Develop in the Absence of a Functional CXCR3 Chemokine Pathway
Next, we explored mechanisms by which CD4+ T cells might accumulate in livers of Tgfb1−/− mice, assessing the CXCR3 pathway. Previously, we showed using gene expression analysis of whole liver RNA that Tgfb1−/− livers exhibit >10-fold upregulation of CXCR3-binding chemokines such as CXCL9.28 Using Luminex analysis of liver lysates, we confirmed at the level of protein that CXCL9 was overexpressed in Tgfb1−/− liver compared with healthy littermate control Tgfb1+/− liver (Figure 3a). Tgfb1−/− livers also exhibited enhanced Cxcr3 mRNA expression (data not shown). To directly test a requirement for CXCR3 in T-cell recruitment and liver damage, Cxcr3−/−Tgfb1−/− double-knockout mice were generated through interbreeding of single knockout mice. Despite the absence of CXCR3, Cxcr3−/−Tgfb1−/− livers exhibited a large CD4+ T-cell lymphocytosis compared with littermate Cxcr3−/−Tgfb1+/− livers; specifically, livers from Cxcr3−/−Tgfb1−/− mice exhibited no difference in either the number (Figure 3b) or percentage (data not shown) of infiltrating CD4+ T cells compared with CXCR3-intact Tgfb1−/− mice. Furthermore, liver injury was readily demonstrable in Cxcr3−/−Tgfb1−/− mice, as measured either by plasma AST (Figure 4) or by histology (Figure 2d). These results demonstrate that CXCR3 is required neither for CD4+ T-cell accumulation nor for subsequent liver damage.
CXCL9 is overexpressed in Tgfb1−/− mouse livers, but CD4+ T-cell accumulation is independent of CXCR3. (a) The CXCR3 ligand CXCL9 was measured in liver lysates isolated from 11-day-old Tgfb1+/− mice and Tgfb1−/− mice and expressed as pg/g wet weight (n=11 for each genotype). Chemokines were measured by Luminex bead assay. Horizontal lines indicate means. The P-value from a statistical comparison of data (Student’s t-test) is shown at the top of the graph. (b) Livers were dissected from 11-day-old mice and weighed. Liver mononuclear cells (MNC) were isolated, counted by hemacytometer, and stained for CD4+ T cells. CD4+ density (cells/g) was calculated. *P<0.01; **P<0.001. NS indicates no significant difference. Mann–Whitney test. n=4–6 mice per genotype. Error bars indicate s.d.
AST elevation in Tgfb1−/− mice is independent of CXCR3 and CCR5. AST was measured from 11-day-old mice of the indicated genotypes. White asterisks reflect statistical differences (*P<0.01; **P<0.001) between Tgfb1−/− mice and their littermate control Tgfb1+/+ mice and Tgfb1+/− mice (aggregated as ‘Tgfb1+’). n=6 or more mice per genotype. For the comparisons indicated by the dashed lines, differences were not significant (NS). Mann–Whitney test. Error bars indicate s.d.
Liver Damage Develops When a Functional CCR5 Chemokine Pathway is Eliminated
We previously showed that several chemokines capable of binding to CCR5 are upregulated greater than 10-fold in Tgfb1−/− liver, compared to heterozygous controls.28 We verified at the protein level that the CCR5-binding chemokines CCL3, CCL4, and CCL5 are significantly overexpressed in Tgfb1−/− liver (Figure 5). Similarly, Tgfb1−/− livers overexpressed mRNA encoding CCR5 (data not shown). Ccr5−/−Tgfb1−/− double-knockout mice were generated through interbreeding of single knockout mice. Ccr5−/−Tgfb1−/− livers developed significant liver damage, indistinguishable from CCR5-intact Tgfb1−/− mice (Figures 4 and 2e).
Chemokine ligands for CCR5 are overexpressed in Tgfb1−/− mouse livers. Chemokine ligands that activate CCR5 were measured in liver lysates isolated from 11-day-old Tgfb1+/− mice and Tgfb1−/− mice and expressed as pg/g wet weight. Each symbol represents one mouse of the indicated genotype. Chemokines were measured by Luminex bead assay. Horizontal lines indicate means. The P-values from statistical comparisons of data (Student’s t-test) are shown at the top of the graphs.
Concurrent Elimination of Both CXCR3 and CCR5 is Permissive for Liver CD4+ T-Cell Accumulation and Hepatocellular Damage, and for the Accumulation of Other Immune Cell Types
To determine whether the two chemokine response pathways are redundant here, we generated Tgfb1−/− mice deficient for both CXCR3 and CCR5. Triple-knockout Cxcr3−/−Ccr5−/−Tgfb1−/− mice nevertheless exhibited robust CD4+ T cell liver lymphocytosis (Figure 6a) as well as acute liver damage (Figures 4 and 2f) equivalent to those observed in CXCR3/CCR5-intact Cxcr3+/+Ccr5+/+Tgfb1−/− mice. In contrast to liver, CD4+ T-cell numbers dropped significantly in spleen, presumably because of their migration into other organs including liver. The reduction in splenic CD4+ T cells was intact in triple-knockout mice (Figure 6b), indicating that CXCR3 and CCR5 are dispensable for this effect.
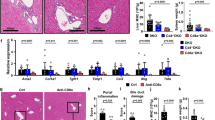
CD4+ T-cell accumulation in Tgfb1−/− mouse liver and reduction in Tgfb1−/− mouse spleen are independent of both CXCR3 and CCR5. (a) Liver and (b) splenic mononuclear cells (MNC) from 11-day-old mice of the indicated genotypes were isolated, counted by hemacytometer, and stained for CD4+ T cells. CD4+ densities (cells/g) were calculated. *P<0.01; **P<0.001; NS indicates no significant difference. Mann–Whitney test. n=4–7 mice per genotype. Error bars indicate s.d.
Next we assessed the accumulation of other immune cell types in liver when both CXCR3 and CCR5 are knocked out in Tgfb1−/− mice. Other cell types, including CD3+ (total T) cells, CD11c+ (dendritic) cells, CD11b+ (myeloid) cells, and F4/80+ (macrophage/Kupffer) cells accumulated in Tgfb1−/− livers compared with Tgfb1+ livers. As observed for CD4+ T cells, the accumulation of these cell types did not require CXCR3/CCR5 (Figure 7).
Accumulation of T cells, dendritic cells, and macrophages in Tgfb1−/− mouse liver is independent of both CXCR3 and CCR5. Liver mononuclear cells (MNC) prepared as in the legend to Figure 6 were stained for the markers CD3, CD11c, CD11b, and F4/80. P-values for statistical comparisons are shown. Increases in some cell populations approached (P<0.10), but did not reach (P<0.05) statistical significance. Error bars indicate s.d.
The Development of Robust type 1 Cytokine/Chemokine Inflammatory Responses in Tgfb1−/− Mice is Independent of CD1d/NKT, CXCR3, and CCR5
Tgfb1−/− mice develop a strong type 1 inflammation with overexpression of Th1-associated cytokines such as IFN-γ. We have previously shown that plasma IFN-γ in Tgfb1−/− mice is derived entirely from the CD4+ T-cell compartment.24 To determine if the over-production of type 1 cytokines is affected by CD1d/NKT or by the chemokine receptors, we measured plasma levels of various cytokines. Plasma from Tgfb1−/− mice had very high levels of IFN-γ, as well as of the Th1-associated cytokines TNF-α and IL-2. High plasma levels of all three cytokines were observed in the double and triple-knockout Tgfb1−/− mouse lines tested (Figure 8), although IL-2 levels were not quite as high in Tgfb1−/− mice lacking CD1d. Thus, the development of a type 1 inflammatory response in Tgfb1−/− mice does not depend on CD1d/NKT, CXCR3, or CCR5.
Overexpression of type 1 cytokines is independent of CD1d/iNKT cells, CXCR3, and CCR5. Plasma samples were isolated at day 11 for mice of the indicated genotypes. Cytokines were measured by Luminex bead assay and results shown as pg/ml. n=3–10 mice per genotype, except for Tgfb1+ mice, for which n=2. Error bars indicate s.e.m.
Next, we assessed chemokine expression levels. As observed for IFN-γ itself, the IFN-γ-induced CXCR3-binding chemokines CXCL9 and CXCL10 were overexpressed in Tgfb1−/− mice, and expression was not dependent on CD1d/NKT, CXCR3, or CCR5. The CC chemokines CCL2, CCL3, CCL4, and CCL5 were also overexpressed to greater or lesser degrees in plasma from Tgfb1−/− mice, and overexpression was largely independent of CD1d/NKT, CXCR3, or CCR5. Closer inspection reveals some interesting interactions. For example, when CXCR3 was absent, CCL2 expression was exacerbated in Tgfb1−/− mice. Such exacerbation was itself CCR5 dependent, as seen in the CXCR3/CCR5 triple-knockout Tgfb1−/− mice. By contrast, in triple-knockout Cxcr3−/−Ccr5−/−Tgfb1−/− mice, but not in double-knockout Cxcr3−/−Tgfb1−/− mice, the expression levels of CCL4 and CCL5 were higher compared with single knockout Tgfb1−/− mice. Interestingly, CXCL2 was not overexpressed in Tgfb1−/− mice, but became so when both CXCR3 and TGF-β1 were knocked out. Finally, there was specificity to overexpression; that is, not all chemokines were overexpressed, as levels of CCL11 (Eotaxin) were not elevated in any of the single, double, or triple Tgfb1−/− knockout mouse lines (Figure 9).
Overexpression of IFN-γ-induced chemokines is independent of CD1d/iNKT cells, CXCR3, and CCR5. Chemokines were analyzed in day 11 samples by Luminex, and results are displayed as in the legend to Figure 8.
Finally, we examined a number of other cytokines. As for IFN-γ, some cytokines (G-CSF, IL-6, IL-10) were strongly overexpressed in Tgfb1−/− mice independent of CD1d/NKT or the chemokine receptors (Figure 10a). Other cytokines (IL-1α, IL-1β, IL-5, M-CSF, IL-12 p40, IL-12 p70, IL-13) were overexpressed to lesser or greater degrees in Tgfb1−/− mice, but expression was exacerbated in Tgfb1−/− mice deficient in CXCR3, but not in Tgfb1−/− mice deficient in both CXCR3 and CCR5 (Figure 10b). Not all cytokines were overexpressed in Tgfb1−/− mice: plasma GM-CSF and IL-15 expression levels were similar in Tgfb1−/− mice and Tgfb1+ mice (Figure 10c), and levels of LIX and of IL-9 were lower in Tgfb1−/− mice (Figure 10d).
Expression levels of specific cytokines are differentially influenced by TGF-β1, CD1d/iNKT cells, CXCR3, and CCR5. Cytokines were analyzed in day 11 samples by Luminex, and results are displayed as in the legend to Figure 8. (a) Some cytokines (G-CSF, IL-6, IL-10) are strongly overexpressed in plasma from Tgfb1−/− mice; expression levels are largely independent of functional Cd1d, Cxcr3, or Ccr5 genes. (b) These cytokines show highest expression when both Tgfb1 and Cxcr3 are knocked out, but levels revert when Ccr5 is additionally knocked out. (c) Expression levels of some cytokines (GM-CSF, IL-15) are unaffected in Tgfb1−/− mice. (d) Expression levels of some cytokines (LIX, IL-9) are reduced in Tgfb1−/− mice.
DISCUSSION
This work demonstrates that T-cell accumulation in liver and the development of liver injury can occur by distinct mechanisms in distinct models of acute CD4+ T-cell-mediated liver inflammation and hepatocellular damage. For example, as opposed to the ConA model of CD4+ T-cell-mediated liver injury, hepatocellular damage in the BALB/c-Tgfb1−/− model does not require iNKT cells. In addition, importantly, a deficiency in both CXCR3 and CCR5 fails to alter CD4+ T-cell accumulation in the liver and subsequent necroinflammatory liver damage. Interestingly, though the ligands for these dominant Th1 chemokine receptors are substantially upregulated in diseased liver, they are not required for CD4+ T-cell migration and accumulation. Thus, at least in this model system, CD4+ T cells can accumulate in inflamed livers independently of either of the traditional chemokine receptors responsible for Th1-cell movement into sites of inflammation.
iNKT cells are required for ConA induced liver damage in mice,11, 12 and it is suggested they have a role in PBC.30 A recent study shows that deficiency of CD1d-restricted iNKT cells can prevent lethality and abrogate inflammation in an ROS-mediated liver damage model.31 Inflammatory innate cell responses, proinflammatory cytokine expression, and hepatocellular death were all substantially decreased in CD1d-deficient mice compared with control mice.31 Similarly, another recent study demonstrated that deficiency of iNKT cells drastically decreased liver injury in a drug-induced liver damage model in mice.32 Several studies support a requirement for iNKT cells in liver injury in the ConA induced model of hepatitis. Interestingly, IL-4 production by iNKT cells is necessary to upregulate granzyme B and FasL, required for liver injury.33 By contrast, in BALB/c-Tgfb1−/− mice the absence of CD1d-restricted iNKT cells did not alter disease parameters, suggesting that a conventional CD4+ T-cell population is responsible for liver damage in this model. Our results are consistent with data from AIH patients, in whom iNKT cells, similar to FoxP3-expressing regulatory T cells, may actually promote tolerance, rather than serve as an effector cell subset in liver damage.34
Cell types other than T cells and iNKT cells can participate in liver damage in acute and chronic liver diseases. Neutrophils are the characteristic cellular component of acute liver injury, are prominent in alcoholic hepatitis, and may also participate in chronic liver diseases.35 Moreover, neutrophils are necessary for the development of liver damage following ConA administration.36 Close inspection of H&E-stained Tgfb1−/− liver sections reveals an abundance of myeloid cells that resemble neutrophils.27 We thus considered the possibility that neutrophils are important for liver damage in Tgfb1−/− mice. To test this hypothesis, we attempted to deplete neutrophils from Tgfb1−/− mice using the myeloid specific monoclonal antibody Gr-1. Despite the successful elimination of >90% of myeloid cells, Gr-1-injected Tgfb1−/− mice nevertheless developed robust liver damage similar to that of unmanipulated Tgfb1−/− mice (JGC and JDG, unpublished data). We conclude that, as for iNKT cells, neutrophils do not contribute to hepatocellular damage in this mouse model of hepatitis.
The involvement of chemokines in Th1-cell migration and accumulation in liver is well described in the literature. In addition, studies have detailed the roles of the inflammatory chemokines CXCR3 and CCR5 as the two dominant receptors for recruitment of Th1 cells into inflamed tissue.37, 38, 39 Moreover, Th1 cells can utilize CXCR3 and/or CCR5 as the chemokine receptors responsible for migration and accumulation in the liver.14 Notably, CXCR3 chemokine levels in HCV patients correlate with disease pathology.19 Another study involving HCV patients proposed preferential T-cell migration into liver architecture based on CCR5 and CXCR3 expression.18 That CD4+ T-cell accumulation in liver in our model is unaffected by CXCR3 and CCR5 deficiency directly shows that other mechanisms can also mediate CD4+ T-cell accumulation in the liver in the setting of autoimmunity. Consistent with this, CXCR3 and CCR5 are not required for CD4+ T-cell accumulation in liver in a virally-induced hepatic steatosis model.40
Mechanisms other than chemokines can participate in the accumulation of T cells in liver parenchyma. Intra-vital microscopy of livers in ConA-treated mice shows that Th1 cells located in the sinusoid or post-sinusoid venules preferentially utilize the integrin α4β1 for adhesion and accumulation in inflamed liver, as antibody neutralization of α4 decreased CD4+ T-cell accumulation in the liver to basal levels.41 Moreover, ConA treatment causes upregulation of VCAM-1 and P-selectin in areas of hepatic blood flow.41 These studies should be interpreted with caution, as they involved in vitro Th1-skewed CD4+ T cells injected via cannulation of the jugular vein. Whether in vitro skewed Th1 cells are phenotypically equivalent to endogenous Th1 cells is uncertain, as is the effect of the injection of a large bolus of T cells (107). Finally, it is notable that significant splenomegaly is observable in BALB/c-Tgfb1−/− mice at postnatal day 11, accompanied by a decreased CD4+ T-cell density in spleen compared with wild-type mice (this study, and Rudner et al23) that may be explained by the migration of CD4+ T cells from spleen to inflamed tissues such as liver. As shown here, the decrease in CD4+ T cells in spleen is independent of CXCR3 and CCR5.
Numerous studies in human liver disease suggest the involvement of specific chemokine pathways in mediating T-cell recruitment to the liver based on correlation of chemokine levels and disease parameters.15, 16, 19 The current study suggests that CD4+ T cells are capable of significant liver accumulation and subsequent damage through additional pathways. While the precise mechanisms responsible for Th1 accumulation in BALB/c-Tgfb1−/− mice are not established, our observations have possibly important implications for the efficacy of chemokine-blocking therapeutic interventions to reduce liver Th1-cell accumulation and hepatocellular damage.
References
Senaldi G, Portmann B, Mowat AP et al. Immunohistochemical features of the portal tract mononuclear cell infiltrate in chronic aggressive hepatitis. Arch Dis Child 1992;67:1447–1453.
Rosen HR . Hepatitis C pathogenesis: mechanisms of viral clearance and liver injury. Liver Transpl. 2003;9:S35–S43.
Grakoui A, Shoukry NH, Woollard DJ et al. HCV persistence and immune evasion in the absence of memory T cell help. Science 2003;302:659–662.
Schlaak JF, Löhr H, Gallati H et al. Analysis of the in vitro cytokine production by liver-infiltrating T cells of patients with autoimmune hepatitis. Clin Exp Immunol 1993;94:168–173.
Rymarchyk SL, Lowenstein H, Mayette J et al. Widespread natural variation in murine natural killer T-cell number and function. Immunology 2008;125:331–343.
Benlagha K, Kyin T, Beavis A et al. A thymic precursor to the NK T cell lineage. Science 2002;296:553–555.
Matsuda JL, Naidenko OV, Gapin L et al. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med 2000;192:741–754.
Gantner F, Leist M, Lohse AW et al. Concanavalin A-induced T-cell-mediated hepatic injury in mice: the role of tumor necrosis factor. Hepatology 1995;21:190–198.
Tiegs G, Hentschel J, Wendel A . A T cell-dependent experimental liver injury in mice inducible by concanavalin A. J Clin Invest 1992;90:196–203.
Knolle PA, Gerken G, Loser E et al. Role of sinusoidal endothelial cells of the liver in concanavalin A-induced hepatic injury in mice. Hepatology 1996;24:824–829.
Toyabe S, Seki S, Iiai T et al. Requirement of IL-4 and liver NK1+ T cells for concanavalin A-induced hepatic injury in mice. J Immunol 1997;159:1537–1542.
Takeda K, Hayakawa Y, Van Kaer L et al. Critical contribution of liver natural killer T cells to a murine model of hepatitis. Proc Natl Acad Sci USA 2000;97:5498–5503.
Osman Y, Kawamura T, Naito T et al. Activation of hepatic NKT cells and subsequent liver injury following administration of alpha-galactosylceramide. Eur J Immunol 2000;30:1919–1928.
Oo YH, Adams DH . The role of chemokines in the recruitment of lymphocytes to the liver. J Autoimmun 2010;34:45–54.
Nishioji K, Okanoue T, Itoh Y et al. Increase of chemokine interferon-inducible protein-10 (IP-10) in the serum of patients with autoimmune liver diseases and increase of its mRNA expression in hepatocytes. Clin Exp Immunol 2001;123:271–279.
Nagayama K, Enomoto N, Miyasaka Y et al. Overexpression of interferon gamma-inducible protein 10 in the liver of patients with type I autoimmune hepatitis identified by suppression subtractive hybridization. Am J Gastroenterol 2001;96:2211–2217.
Honda M, Kawai H, Shirota Y et al. cDNA microarray analysis of autoimmune hepatitis, primary biliary cirrhosis and consecutive disease manifestation. J Autoimmun 2005;25:133–140.
Shields PL, Morland CM, Salmon M et al. Chemokine and chemokine receptor interactions provide a mechanism for selective T cell recruitment to specific liver compartments within hepatitis C-infected liver. J Immunol 1999;163:6236–6243.
Zeremski M, Petrovic LM, Chiriboga L et al. Intrahepatic levels of CXCR3-associated chemokines correlate with liver inflammation and fibrosis in chronic hepatitis C. Hepatology 2008;48:1440–1450.
Affo S, Bataller R . RANTES antagonism: a promising approach to treat chronic liver diseases. J Hepatol 2011;55:936–938.
Li MO, Wan YY, Sanjabi S et al. Transforming growth factor-beta regulation of immune responses. Annu Rev Immunol 2006;24:99–146.
Gorham JD, Lin JT, Sung JL et al. Genetic regulation of autoimmune disease: BALB/c background TGF-beta 1- deficient mice develop necroinflammatory IFN-gamma-dependent hepatitis. J Immunol 2001;166:6413–6422.
Rudner LA, Lin JT, Park IK et al. Necroinflammatory liver disease in BALB/c background, TGF-beta 1-deficient mice requires CD4+ T cells. J Immunol 2003;170:4785–4792.
Robinson RT, Wang J, Cripps JG et al. End-organ damage in a mouse model of fulminant liver inflammation requires CD4+ T cell production of IFN-gamma but is independent of Fas. J Immunol 2009;182:3278–3284.
Hancock WW, Lu B, Gao W et al. Requirement of the chemokine receptor CXCR3 for acute allograft rejection. J Exp Med 2000;192:1515–1520.
Pan J, Burdick MD, Belperio JA et al. CXCR3/CXCR3 ligand biological axis impairs RENCA tumor growth by a mechanism of immunoangiostasis. J Immunol 2006;176:1456–1464.
Cripps JG, Wang J, Maria A et al. Type 1 T helper cells induce the accumulation of myeloid-derived suppressor cells in the inflamed Tgfb1 knockout mouse liver. Hepatology 2010;52:1350–1359.
Milks MW, Cripps JG, Lin H et al. The role of Ifng in alterations in liver gene expression in a mouse model of fulminant autoimmune hepatitis. Liver Int 2009;29:1307–1315.
Mendiratta SK, Martin WD, Hong S et al. CD1d1 mutant mice are deficient in natural T cells that promptly produce IL-4. Immunity 1997;6:469–477.
Kita H, Naidenko OV, Kronenberg M et al. Quantitation and phenotypic analysis of natural killer T cells in primary biliary cirrhosis using a human CD1d tetramer. Gastroenterology 2002;123:1031–1043.
Ishikawa S, Ikejima K, Yamagata H et al. CD1d-restricted natural killer T cells contribute to hepatic inflammation and fibrogenesis in mice. J Hepatol 2011;54:1195–1204.
Cheng L, You Q, Yin H et al. Involvement of natural killer T cells in halothane-induced liver injury in mice. Biochem Pharmacol 2010;80:255–261.
Kaneko Y, Harada M, Kawano T et al. Augmentation of Valpha14 NKT cell-mediated cytotoxicity by interleukin 4 in an autocrine mechanism resulting in the development of concanavalin A-induced hepatitis. J Exp Med 2000;191:105–114.
Ferri S, Longhi MS, De Molo C et al. A multifaceted imbalance of T cells with regulatory function characterizes type 1 autoimmune hepatitis. Hepatology 2010;52:999–1007.
Ramadori G, Saile B . Inflammation, damage repair, immune cells, and liver fibrosis: specific or nonspecific, this is the question. Gastroenterology 2004;127:997–1000.
Bonder CS, Ajuebor MN, Zbytnuik LD et al. Essential role for neutrophil recruitment to the liver in concanavalin A-induced hepatitis. J Immunol 2004;172:45–53.
Sallusto F, Lanzavecchia A, Mackay CR . Chemokines and chemokine receptors in T-cell priming and Th1/Th2-mediated responses. Immunol Today 1998;19:568–574.
Loetscher P, Uguccioni M, Bordoli L et al. CCR5 is characteristic of Th1 lymphocytes. Nature 1998;391:344–345.
Qin S, Rottman JB, Myers P et al. The chemokine receptors CXCR3 and CCR5 mark subsets of T cells associated with certain inflammatory reactions. J Clin Invest 1998;101:746–754.
Holst PJ, Orskov C, Qvortrup K et al. CCR5 and CXCR3 are dispensable for liver infiltration, but CCR5 protects against virus-induced T-cell-mediated hepatic steatosis. J Virol 2007;81:10101–10112.
Bonder CS, Norman MU, Swain MG et al. Rules of recruitment for Th1 and Th2 lymphocytes in inflamed liver: a role for alpha-4 integrin and vascular adhesion protein-1. Immunity 2005;23:153–163.
Acknowledgements
This work was supported by National Institutes of Health Grants AI078195 (JDG) and P20RR16437 from the COBRE Program of the National Center for Research Resources, as well as by a grant from the Hitchcock Foundation (JDG). JGC was supported by National Institutes of Health Training Grant T32AI07363. We thank Beverly Gorham and Christine Kretowicz for assistance in mouse breeding and screening.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
In autoimmune hepatitis and viral hepatitis, liver damage is dependent on infiltration by CD4+ T cells, which can accumulate independently of either of the traditional chemokine receptors responsible for Th1-cell movement into sites of inflammation. CD4+ T cells are therefore capable of significant liver accumulation and subsequent damage through additional unidentified pathways.
Rights and permissions
About this article
Cite this article
Cripps, J., Celaj, S., Burdick, M. et al. Liver inflammation in a mouse model of Th1 hepatitis despite the absence of invariant NKT cells or the Th1 chemokine receptors CXCR3 and CCR5. Lab Invest 92, 1461–1471 (2012). https://doi.org/10.1038/labinvest.2012.104
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2012.104
Keywords
This article is cited by
-
Regulatory T Cell Plasticity and Stability and Autoimmune Diseases
Clinical Reviews in Allergy & Immunology (2020)