Abstract
The biological roles of microRNAs (miRNAs) have been extensively studied. miRNA122 represents more than half of the miRNAs expressed in the liver and has various physiological and pathological functions, which include enhancing hepatitis virus replication, regulating lipid metabolism and suppressing hepatocellular carcinoma. miRNAs, whether globally or individually, have been linked with hepatocarcinogenesis. Furthermore, some miRNAs have been shown to be involved in the pathogenesis of nonalcoholic steatohepatitis. Using nucleotide-based strategies, these miRNAs may be developed as potential therapeutic targets. Because changes in miRNA expression can be measured in sera, they may be used as non-invasive biomarkers if they correctly reflect the pathological state of the liver. In this review, we show the biological roles of representative miRNAs in liver disease and discuss the current issues that remain to be clarified for future clinical applications.
Similar content being viewed by others
Introduction
MicroRNAs (miRNAs) are short, single-stranded, noncoding RNAs that are expressed in most organisms.1 As the discovery of the miRNA lin-4 in Caenorhabditis elegans,2, 3 1881 miRNA precursors and 2588 mature miRNA human sequences have been deposited in miRBase, a public repository hosted by the Sanger Institute, as of June 2014.4 It is speculated that miRNAs regulate >30% of protein-coding genes in humans.5, 6, 7 By regulating gene expression, miRNAs are involved in various physiological and pathological processes.8, 9 In particular, a growing number of studies have been published, especially since 2005, connecting miRNAs with liver disease. There are now >3000 published reports. Because it is impossible to summarize all of the results here, we describe the roles of representative miRNAs that are considered important in the pathology of liver disease.
Biological roles of miR122 in liver disease
miRNA expression is tissue specific. miR122 is an miRNA that makes up more than half of all of the miRNAs expressed in the liver, followed by miR192 and miR199a/b-3p, accounting for 52%, 17% and 5%, respectively.10 Therefore, miR122 is thought to have an important role in the liver. The most striking feature of miR122 is its positive role in hepatitis C virus (HCV) replication. The 5′ internal ribosome entry site of the HCV genome contains two possible miR122-binding sites that are essential for efficient HCV replication in the liver.11 miR122 interacts with these sites and enhances replication of HCV, at least in part by enhancing viral translation. Eight years after the first report of the positive effects of miR122 on HCV replication,11 miravirsen, an locked nucleic acid, antagomir against miR122 that suppresses HCV replication, was developed as the first patient drug based on miRNAs. In a phase 2 clinical trial, miravirsen was well tolerated and produced a good virological response.12 Thus, miRNA-based therapeutics may change the current treatment of some diseases.
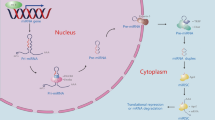
A more recent study reported the outcome of the interaction between miR122 and HCV. During infection, HCV RNA specifically sequesters miR122, impairing the intrinsic function of miR122 and derepressing normal miR122 targets. This ‘miRNA sponge’ effect was originally reported with the phosphatase and tensin homolog pseudogene 1 (PTENP1), which harbors the same miRNA target sequences as phosphatase and tensin homolog deleted from chromosome 10 (PTEN).13 Normal expression of PTENP1 may work as a decoy for several miRNAs, which also target PTEN. Once PTENP1 expression is lost, the decoy effects are also lost, resulting in further PTEN targeting and a resulting decrease in its expression (Figure 1). This previously unidentified and surprising decoy function of the pseudogene may have a tumor-suppressive role by regulating miRNA function.13 The report convincingly showed that, similar to PTENP1, HCV RNA acts as a decoy for miR122 and impairs its normal function (Figure 2), which may facilitate the long-term oncogenic potential of HCV.14
PTEN is protected from microRNA binding by PTENP1. PTENP1 (upper) and PTEN (lower) 3′ UTRs contain a highly conserved domain. miRNAs targeting the PTEN 3′ UTR in this domain can be trapped by the domain in the PTENP1 3′ UTR in the presence of PTENP1. Through these functional interactions, PTENP1 is biologically active as it can regulate cellular levels of PTEN by trapping miRNAs. miRNA, microRNA; PTEN, phosphatase and tensin homolog deleted from chromosome 10; PTENP1, phosphatase and tensin homolog pseudogene 1; UTRs, untranslated regions. A full color version of this figure is available at the Journal of Human Genetics journal online.
HCV RNA sequesters miR122 and derepresses miR122-targeting genes. (a) miR122 normally suppresses the expression levels of its target genes, which contain miR122-targeting sequences in their 3′ UTRs. Solid lines in 3′ UTR indicate the positions of possible miR122 target sequences. (b) In HCV-infected cells, viral RNA specifically sequesters miR122 by its miR122 target sequences in its 5′ IRES (indicated by solid lines in 5′ IRES) to derepress its normal host target genes. These may be responsible for the pathogenesis of long-term HCV infection. HCV, hepatitis C virus; IRES, internal ribosome entry site; UTRs, untranslated regions. A full color version of this figure is available at the Journal of Human Genetics journal online.
To determine the biological role of miR122, miR122-deleted mice were generated. Deletion of mouse miR122 caused the mice to develop steatohepatitis and subsequent liver tumors.15, 16 Serum levels of cholesterol and triglyceride were also significantly reduced in these mice. There results are consistent with previous studies using miR122 inhibitors in vivo.17, 18 miR122 is the first reported miRNA involved in lipid metabolism. Several studies have reported that antagonism of miR122 results in a sustained decrease in plasma cholesterol levels in mice, non-human primates and humans.12, 17, 18, 19, 20 Despite these results, the molecular mechanisms underlying miR122-regulated cholesterol levels still remain to be elucidated. Although many genes associated with cholesterol metabolism are affected by miR122 inhibition, they are not always direct targets of miR122.21 Because miR122-deleted mice showed both steatohepatitis and reduced serum levels of lipid, miR122 may target genes important in lipoprotein secretion.21 To address the precise role of miR122 in lipid metabolism, comprehensive miR122 target profiling and functional analyses are important.
miR122 also acts as a tumor suppressor. This tumorigenic role was confirmed with the loss of miR122 in knockout mice, as described above.15, 16 The expression of miR122 is decreased in half of the cases of hepatocellular carcinoma (HCC), particularly non-viral HCC.10 Decreased expression of miR122 is also closely linked with aggressiveness and chemo-resistance of HCC.22, 23 Therefore, miR122 seems to have an important role as a tumor suppressor in the liver.
Although the details are not described here, miR122 is also involved in other hepatic functions and disease, including proper liver development,24, 25 hepatitis B virus pathogenesis,26 circadian rhythms,27 iron metabolism28 and others.24, 25 Thus, miR122, the major miRNA in the liver, is involved in various kinds of physiological functions in addition to liver disease (Figure 3).
miR122 has many functions in the liver. miR122 is involved in HCV replication, lipid metabolism, hepatocarcinogenesis and cell differentiation, which are pathologically and physiologically important in the liver. HCV, hepatitis C virus. A full color version of this figure is available at the Journal of Human Genetics journal online.
Biogenesis and functions of miRNAs
Although individual miRNAs may have important roles in the liver, it is also possible that almost all miRNAs are deregulated because of impaired miRNA biogenesis. Initially, primary miRNAs, which contain stem–loop structures, are transcribed by RNA polymerase II.8 These primary miRNAs are processed by a microprocessor complex composed of Drosha (RNAase III)29 and DiGeorge syndrome critical region gene 8/partner of Drosha30 in the nucleus.31 The processed products are ~65-nucleotide hairpin-shaped precursors (pre-miRNAs) that are transported to the cytoplasm via exportin-5.32, 33 Pre-miRNAs are further cleaved into mature miRNAs by Drosha and Dicer RNA polymerase III. Mature miRNA duplexes are loaded onto an RNA-induced silencing complex and are unwound into the single-stranded mature form.34, 35, 36 The resulting co-complex directly targets the 3′-untranslated regions of target mRNAs, depending on sequence similarities, to negatively regulate their expression by enhancing mRNA cleavage or inhibiting translation.8, 37
Given that Dicer is essential for most miRNA processing, conditional mice with deletion of the Dicer gene in hepatocytes may provide a useful model for determining the physiological function of global miRNAs in the liver.38 In these mutant mice, liver zonation was deregulated with diffuse expression of periportal proteins, such as carboxykinase and E-cadherin.38 Dicer-deficient hepatocytes result in prominent steatosis and depletion of glycogen stores. Mutant mice developed HCC after 1 year of age with Dicer-deficient hepatocytes.38 These results suggest that Dicer and/or global miRNAs in the liver have vital roles in the regulation of the liver development and metabolism, and in the suppression of tumor development.38 Consistent with this, one study reported that low dicer expression in human HCC is linked with a poor prognosis.39 Thus, it may be important to consider not only individual miRNA function, but also gross miRNA function in light of the pathogenesis of liver disease.
miRNAs and HCC
Numerous reports have described the deregulated expression of miRNAs in human HCC. Most studies have selected candidate miRNA(s) and revealed their target genes, which may be involved in carcinogenesis, and compared miRNA expression levels between cancerous and non-tumorous background tissues. The list of miRNAs with deregulated miRNA expression has already been described in several excellent reviews.40, 41 Here we describe select miRNAs that may have crucial roles in HCC.
The expression level of miR26 has been linked to the prognosis of HCC.42 Patients with higher miR26 expression show better prognosis following surgery. Consistent with results from a clinical study, miR26 has been confirmed as a tumor suppressor in mice. In a c-Myc-driven hepatocarcinogenesis model in mice, miR26 delivery by adeno-associated virus significantly suppressed tumor burden.43 Thus, miR26 expression suppresses HCC, and therefore may be useful for the prevention or treatment of HCC.
Another well-known tumor-suppressive miRNA is let-7. The oncogenic and RNA-binding proteins LIN28 and LIN28B specifically block the processing of let-7 precursors into mature miRNAs,44 suggesting that their overexpression might promote malignancy by repressing let-7. On the other hand, let-7 targets LIN28 and LIN28B, decreasing their protein levels. Thus, LIN28 and let-7 antagonize one other under normal physiological conditions. If these antagonizing effects are deregulated, pathological conditions may occur (Figure 4).
Deregulated balance between LIN28/28B and let-7 leads to oncogenesis. LIN28/28B and let-7 antagonize each other under normal physiological conditions. Once LIN28/28B expression is enhanced, for example, by inflammation and the balance collapses, LIN28/28B expression is augmented and let-7 expression further suppressed, resulting in the oncogenic effects of LIN28/28B. A full color version of this figure is available at the Journal of Human Genetics journal online.
Inflammation is considered a major cause of cancer.45, 46 HCC frequently occurs in persistently inflamed liver tissues that are a result of chronic hepatitis viral infection or nonalcoholic steatohepatitis (NASH). However, the molecular linkage between chronic inflammation and carcinogenesis has not been well characterized. miRNAs may be involved in chronic inflammation-induced carcinogenesis. In fact, several studies have clarified one such linkage, in which miRNAs may serve as a bridge between continuous inflammation and carcinogenesis. One report in particular described a positive-feedback loop in an inflammatory response mediated by NF-κB that activates Lin28B transcription (Figure 5).47 LIN28B, an inhibitor of the pre-let-7 processing described above, reduces mature let-7 levels. Let-7 normally inhibits interleukin 6 (IL-6) expression, resulting in higher levels of IL-6 than usually achieved by NF-κB activation. IL-6-mediated signal transducer and activator of transcription 3 (STAT3) activation is necessary for transformation and IL-6 activates NF-κB, completing the positive-feedback loop. Although the experiments in this study primarily used the breast cancer cell line MCF10A, a similar feedback loop was observed in HCC tissues. The authors termed these mechanisms an ‘epigenetic switch’ because the loop maintains the epigenetic transformed state even in the absence of inflammation.
A model bridging chronic inflammation and transformation by miRNA. Inflammation triggers activation of NF-κB, which leads to the transcription of LIN28B. LIN28B inhibits the production of let-7, which normally inhibits IL-6 expression, resulting in higher levels of IL-6 than usually achieved by NF-κB activation. IL-6-mediated STAT3 activation is necessary for transformation and IL-6 activates NF-κB, completing the positive-feedback loop. IL-6, interleukin 6; miRNA, microRNA. A full color version of this figure is available at the Journal of Human Genetics journal online.
Another paper reported a similar but distinct observation: when using diethyl nitrosamine-induced foci of altered hepatocytes (FAH), LIN28-expressing cells are present in FAH, in which let-7 is downregulated, resulting in the enhanced expression of IL-6, mediating the progression of malignancies from progenitors. An important difference between the cells in FAH and those in early hepatocarcinogenesis is that IL-6 signaling is autocrine, mediated by reduced let-7 due to the upregulation of LIN28B in FAH cells. This mechanism may contribute to malignant progression from HCC progenitor cells.48 Although these hypotheses may be interesting, their relevance in hepatocarcinogenesis needs to be further confirmed to determine the applicability and reproducibility of these findings.
miRNAs and NASH
Deregulated miRNAs may also be involved in the pathogenesis of NASH.49 The list of miRNAs with deregulated expression in NASH is described in several excellent reviews.49, 50 Here we describe miR21 and miR103/107.
miR21 levels are upregulated in the liver of NASH patients compared with control subjects or simple steatosis patients.51 Low-density lipoprotein receptor (LDLR)-deleted mice (Ldlr −/−) fed a high fat diet show aggressive liver inflammation, which can be used as an NASH model.51 miR21 expression in the livers of these mice is also increased. Pharmacological inhibition of miR21 using antagomir-2 reduced hepatic inflammation and liver fibrosis, suggesting that miR21 has a central role in liver inflammation induced by steatosis.51 Hence, antagomir-21 may be useful as a therapeutic option against NASH. Importantly, miR21 is expressed mainly in inflammatory and biliary cells, and not in hepatocytes. Although peroxisome proliferator-activated receptor alpha was raised as a possible target in this study, the precise mechanisms regarding the role of miR21 in the pathogenesis of NASH remain to be determined.
Another landmark study about miRNA and NASH reported that miR103/107 expression is upregulated in the liver of alcoholic steatohepatitis and NASH patients.52 Although the sequences of mature miR103 and 107 differ by one nucleotide at position 21 and cannot be discriminated by northern blotting, both miRNAs were upregulated in the liver of alcoholic steatohepatitis and NASH.52 Overexpression of these miRNAs induces glucose intolerance, and, conversely, silencing by antagomirs results in improved glucose tolerance and insulin sensitivity in ob/ob obese mice.52 These results suggest that miR103 and 107 regulate insulin sensitivity.52 A transgenic model that constitutively overexpresses miR103 exhibited glucose intolerance. Apigenin, a flavonoid that inhibits the maturation of miRNAs, improved the pathogenic status.53 These results suggest that miR103 may be a promising therapeutic target and the modulation of its expression may be a good candidate for improving insulin sensitivity.
Circulating miRNAs as diagnostic markers
miRNAs in the circulation are relatively stable.54 Non-coding RNAs, including miRNAs, are passively released from dying cells or actively released by inclusion in an exosome or in microvesicles, or in a protein-bound form. Microarrays, PCR methods and next-generation sequencing technologies are currently being used to examine the levels of circulating noncoding RNAs. Easily assessable serum-based miRNAs may be useful as novel biomarkers for diagnostic or prognostic purposes in various diseases.
Utilizing a single or several circulating miRNAs as diagnostic markers for HCC has been extensively reported, although the specificity is relatively poor.55 A combination of multiple circulating miRNAs instead of a single miRNA may offer more specificity and sensitivity as a diagnostic marker for HCC diagnosis and prognosis prediction. For example, a panel consisting of seven miRNAs (miR29a, 29c, miR133a, miR143, miR192 and miR505) was recently reported to have higher diagnostic accuracy of HCC (area under the curve=0.833 to detect a smaller size of HCC) than AFP, a commonly used marker, in hepatitis B virus-positive patients.56 Although these results are promising, several other panels using different miRNA sets have also been investigated for similar diagnostic purposes with high accuracy. To confirm the reproducibility and ensure the results are useful for a clinical setting, standardized protocols and well-designed prospective studies will be essential in the future.
Similar to the cases of HCC diagnosis, some miRNAs have been tested for the diagnosis of NASH. For example, 84 circulating miRNAs were assessed to determine if they are differentially expressed in patients with nonalcoholic fatty liver disease.57 Among them, miR122 circulating levels were most markedly changed (7.2-fold change in NASH versus controls, and 3.1-fold change in NASH versus simple steatosis). miR122 serum levels were significantly associated with the nonalcoholic fatty liver disease activity score (NAS). The area under the curve, which indicates the ability of miR122 to discriminate an advanced disease from the mild form, was about 0.7. This is fair, but more sensitive and specific markers are needed. Panels using several miRNAs, similar to the cases in HCC diagnosis, may also be necessary for NASH diagnosis. Although the development of non-invasive diagnostic methods for NASH is promising, more work is still needed to improve the reliability of circulating noncoding RNAs as novel biomarkers.
miRNAs as novel therapeutics against liver disease
Given the mounting evidence that miRNAs are frequently deregulated in HCC and may be involved in oncogenesis, these miRNAs may provide novel molecular targets for therapeutic intervention. However, due to the complexity associated with pleiotropic miRNA functions, the number of clinical trials has been limited.58 The leading nucleotide-targeting therapy, miravirsen, an locked nucleic acid-based anti-miR122 against HCV replication, has been successful in a phase IIa study.12 In addition, MRX34, a liposome-formulated miR-34 mimic developed by Mirna Therapeutics (Austin, TX, USA), produced complete HCC regression in mouse models,59 and a phase I study is currently recruiting patients with advanced liver cancer for HCC therapeutic intervention (NCT01829971). Regulus Therapeutics (San Diego, CA, USA) is developing anti-miR122 (RG-101) and anti-miR103 (RG-125) for anti-HCV and anti-NASH, respectively. Enrollment of patients in phase II has been completed for anti-miR122 and enrollment of patients in phase I is about to start for anti-miR103 with AstraZeneca (London, UK). On the basis of the results that anti-miR21 and anti-miR221 prolonged survival time in a preclinical mouse model that genetically develops HCC, Regulus Therapeutics is also developing anti-miRNAs against these targets for clinical use. In addition, a miR7 mimic is currently being developed by MiReven (Nedians, WA, Australia) to target HCC. Mir7 targets the phosphoinositide 3-kinase pathway and has been shown to decrease tumor growth both in vitro and in vivo.60
Challenges for clinical translation
The biological roles of miRNAs have been extensively studied. However, the reproducibility of reported results are not satisfactory at present, possibly because miRNAs associate with multiple targets given the ambiguity in their sequence matches. The fact that miRNAs have multiple targets and the fact that mRNAs are targeted by multiple miRNAs further complicates the interpretation of any results. More solid data using sophisticated models such as gene-modified mice or well-controlled clinical studies are needed. System biology approaches will also be essential to fully understand the biological roles of miRNAs. Additional issues that should be considered before better clinical translation are as follows:
-
1
For the development of useful biomarkers using circulating miRNAs, specificity and sensitivity, as well as methods to measure small amounts of RNA in sera, with high reproducibility and the universal control to adjust data from different times and samples will be necessary.
-
2
For the development of effective therapeutics using miRNAs, identification of the miRNAs important in pathogenesis will be critical, although passive miRNAs may be utilized as prognostic and diagnostic markers.
-
3
For the development of effective interventions using miRNA-related oligonucleotides, the delivery method, improved oligonucleotide modification and safety must be considered further. Because miRNAs generally have diverse effects because they target multiple mRNAs, undesired outcomes or so called ‘off-target effects,’ may be encountered, even when a specific miRNA is targeted.
Conclusions
The discovery of miRNA has, without doubt, opened up new possibilities for understanding the molecular mechanisms of gene regulation. The speed of discovery in this field is astonishing. In fact, novel therapeutics targeting miRNAs have already been successfully tested in clinical trials. Some miRNAs may also be clinically useful as novel biomarkers. In addition, the discovery of novel concepts in the pathogenesis of hepatocarcinogenesis frequently involves miRNA. However, several important issues remain to be resolved. Continuous research will be essential for the development of truly innovative concepts in our understanding of pathogenesis and its connection with miRNAs and to transform the obtained knowledge into real clinical applications.
References
Carrington, J. & Ambros, V. Role of microRNAs in plant and animal development. Science 301, 336–338 (2003).
Lee, R. C., Feinbaum, R. L. & Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 (1993).
Wightman, B., Ha, I. & Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862 (1993).
Kozomara, A. & Griffiths-Jones, S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39, D152–D157 (2011).
John, B., Enright, A. J., Aravin, A., Tuschl, T., Sander, C. & Marks, D. S. Human MicroRNA targets. PLoS Biol. 2, e363 (2004).
Krek, A., Grün, D., Poy, M. N., Wolf, R., Rosenberg, L., Epstein, E. J. et al. Combinatorial microRNA target predictions. Nat. Genet. 37, 495–500 (2005).
Lewis, B. P., Burge, C. B. & Bartel, D. P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 (2005).
Ambros, V. The functions of animal microRNAs. Nature 431, 350–355 (2004).
Bartel, D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004).
Hou, J., Lin, L., Zhou, W., Wang, Z., Ding, G., Dong, Q. et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell 19, 232–243 (2011).
Jopling, C. L., Yi, M., Lancaster, A. M., Lemon, S. M. & Sarnow, P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 309, 1577–1581 (2005).
Janssen, H. L., Reesink, H. W., Lawitz, E. J., Zeuzem, S., Rodriguez-Torres, M., Patel, K. et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 368, 1685–1694 (2013).
Poliseno, L., Salmena, L., Zhang, J., Carver, B., Haveman, W. J. & Pandolfi, P. P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465, 1033–1038 (2010).
Luna, J. M., Scheel, T. K., Danino, T., Shaw, K. S., Mele, A., Fak, J. J. et al. Hepatitis C virus RNA functionally sequesters miR-122. Cell 160, 1099–1110 (2015).
Tsai, W. C., Hsu, S. D., Hsu, C. S., Lai, T. C., Chen, S. J., Shen, R. et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J. Clin. Invest. 122, 2884–2897 (2012).
Hsu, S. H., Wang, B., Kota, J., Yu, J., Costinean, S., Kutay, H. et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J. Clin. Invest. 122, 2871–2883 (2012).
Elmen, J., Lindow, M., Schutz, S., Lawrence, M., Petri, A., Obad, S. et al. LNA-mediated microRNA silencing in non-human primates. Nature 452, 896–899 (2008).
Lanford, R. E., Hildebrandt-Eriksen, E. S., Petri, A., Persson, R., Lindow, M., Munk, M. E. et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327, 198–201 (2010).
Esau, C., Davis, S., Murray, S. F., Yu, X. X., Pandey, S. K., Pear, M. et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 3, 87–98 (2006).
Krutzfeldt, J., Rajewsky, N., Braich, R., Rajeev, K., Tuschl, T., Manoharan, M. et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438, 685–689 (2005).
Lewis, A. P. & Jopling, C. L. Regulation and biological function of the liver-specific miR-122. Biochem. Soc. Trans. 38, 1553–1557 (2010).
Kojima, K., Takata, A., Vadnais, C., Otsuka, M., Yoshikawa, T., Akanuma, M. et al. MicroRNA122 is a key regulator of alpha-fetoprotein expression and influences the aggressiveness of hepatocellular carcinoma. Nat. Commun. 2, 338 (2011).
Kishikawa, T., Otsuka, M., Ohno, M., Yoshikawa, T., Takata, A. & Koike, K. Circulating RNAs as new biomarkers for detecting pancreatic cancer. World J. Gastroenterol. 21, 8527–8540 (2015).
Xu, H., He, J., Xiao, Z., Zhang, Q., Chen, Y., Zhou, H. et al. Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. Hepatology 52, 1431–1442 (2010).
Laudadio, I., Manfroid, I., Achouri, Y., Schmidt, D., Wilson, M. D., Cordi, S. et al. A feedback loop between the liver-enriched transcription factor network and miR-122 controls hepatocyte differentiation. Gastroenterology 142, 119–129 (2012).
Wang, X. W., Heegaard, N. H. & Orum, H. MicroRNAs in liver disease. Gastroenterology 142, 1431–1443 (2012).
Gatfield, D., Le Martelot, G., Vejnar, C., Gerlach, D., Schaad, O., Fleury-Olela, F. et al. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev. 23, 1313–1326 (2009).
Castoldi, M., Vujic Spasic, M., Altamura, S., Elmén, J., Lindow, M., Kiss, J. et al. The liver-specific microRNA miR-122 controls systemic iron homeostasis in mice. J. Clin. Invest. 121, 1386–1396 (2011).
Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J. et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415–419 (2003).
Han, J., Lee, Y., Yeom, K. H., Kim, Y. K., Jin, H. & Kim, V. N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 18, 3016–3027 (2004).
Denli, A. M., Tops, B. B., Plasterk, R. H., Ketting, R. F. & Hannon, G. J. Processing of primary microRNAs by the Microprocessor complex. Nature 432, 231–235 (2004).
Yi, R., Qin, Y., Macara, I. G. & Cullen, B. R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17, 3011–3016 (2003).
Lund, E., Güttinger, S., Calado, A., Dahlberg, J. E. & Kutay, U. Nuclear export of microRNA precursors. Science 303, 95–98 (2004).
Maniataki, E. & Mourelatos, Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 19, 2979–2990 (2005).
Mourelatos, Z., Dostie, J., Paushkin, S., Sharma, A., Charroux, B., Abel, L. et al. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 16, 720–728 (2002).
Gregory, R. I., Chendrimada, T. P., Cooch, N. & Shiekhattar, R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123, 631–640 (2005).
Bartel, D. P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 (2009).
Sekine, S., Ogawa, R., Ito, R., Hiraoka, N., McManus, M. T., Kanai, Y. et al. Disruption of Dicer1 induces dysregulated fetal gene expression and promotes hepatocarcinogenesis. Gastroenterology 136, 2304–2315. e1–e4. (2009).
Kitagawa, N., Ojima, H., Shirakihara, T., Shimizu, H., Kokubu, A., Urushidate, T. et al. Downregulation of the microRNA biogenesis components and its association with poor prognosis in hepatocellular carcinoma. Cancer Sci. 104, 543–551 (2013).
Otsuka, M., Kishikawa, T., Yoshikawa, T., Ohno, M., Takata, A., Shibata, C. et al. The role of microRNAs in hepatocarcinogenesis: current knowledge and future prospects. J. Gastroenterol. 49, 173–184 (2014).
Wu, Q., Liu, H. O., Liu, Y. D., Liu, W. S., Pan, D., Zhang, W. J. et al. Decreased expression of hepatocyte nuclear factor 4α (Hnf4α)/MicroRNA-122 (miR-122) axis in hepatitis B virus-associated hepatocellular carcinoma enhances potential oncogenic GALNT10 protein activity. J. Biol. Chem. 290, 1170–1185 (2015).
Ji, J., Shi, J., Budhu, A., Yu, Z., Forgues, M., Roessler, S. et al. MicroRNA expression, survival, and response to interferon in liver cancer. N. Engl. J. Med. 361, 1437–1447 (2009).
Kota, J., Chivukula, R. R., O’Donnell, K. A., Wentzel, E. A., Montgomery, C. L., Hwang, H. W. et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 137, 1005–1017 (2009).
Viswanathan, S. R., Daley, G. Q. & Gregory, R. I. Selective blockade of microRNA processing by Lin28. Science 320, 97–100 (2008).
Clevers, H. At the crossroads of inflammation and cancer. Cell 118, 671–674 (2004).
Grivennikov, S. I., Greten, F. R. & Karin, M. Immunity, inflammation, and cancer. Cell 140, 883–899 (2010).
Iliopoulos, D., Hirsch, H. A. & Struhl, K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 139, 693–706 (2009).
He, G. et al. Identification of Liver Cancer Progenitors Whose Malignant Progression Depends on Autocrine IL-6 Signaling. Cell 155, 384–396 (2013).
Zarfeshani, A., Ngo, S. & Sheppard, A. M. MicroRNA Expression Relating to Dietary-Induced Liver Steatosis and NASH. J. Clin. Med. 4, 1938–1950 (2015).
Szabo, G. & Csak, T. Role of MicroRNAs in NAFLD/NASH. Dig. Dis. Sci. 61, 1314–1324 (2016).
Loyer, X., Paradis, V., Hénique, C., Vion, A. C., Colnot, N., Guerin, C. L. et al. Liver microRNA-21 is overexpressed in non-alcoholic steatohepatitis and contributes to the disease in experimental models by inhibiting PPARα expression. Gut. doi:10.1136/gutjnl-2014-308883 (2015).
Trajkovski, M., Hausser, J., Soutschek, J., Bhat, B., Akin, A., Zavolan, M. et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 474, 649–653 (2011).
Ohno, M., Shibata, C., Kishikawa, T., Yoshikawa, T., Takata, A., Kojima, K. et al. The flavonoid apigenin improves glucose tolerance through inhibition of microRNA maturation in miRNA103 transgenic mice. Sci. Rep. 3, 2553 (2013).
Mitchell, P. S., Parkin, R. K., Kroh, E. M., Fritz, B. R., Wyman, S. K., Pogosova-Agadjanyan, E. L. et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl Acad. Sci. USA 105, 10513–10518 (2008).
Zhang, Y. C., Xu, Z., Zhang, T. F. & Wang, Y. L. Circulating microRNAs as diagnostic and prognostic tools for hepatocellular carcinoma. World J. Gastroenterol. 21, 9853–9862 (2015).
Lin, X. J., Chong, Y., Guo, Z. W., Xie, C., Yang, X. J., Zhang, Q. et al. A serum microRNA classifier for early detection of hepatocellular carcinoma: a multicentre, retrospective, longitudinal biomarker identification study with a nested case-control study. Lancet Oncol. 16, 804–815 (2015).
Pirola, C. J., Fernández Gianotti, T., Castaño, G. O., Mallardi, P., San Martino, J., Mora Gonzalez Lopez Ledesma, M. et al. Circulating microRNA signature in non-alcoholic fatty liver disease: from serum non-coding RNAs to liver histology and disease pathogenesis. Gut. 64, 800–812 (2015).
Shibata, C., Otsuka, M., Kishikawa, T., Yoshikawa, T., Ohno, M., Takata, A. et al. Current status of miRNA-targeting therapeutics and preclinical studies against gastroenterological carcinoma. Mol. Cell. Ther. 1, 5 (2013).
Xu, Y., Liu, L., Liu, J., Zhang, Y., Zhu, J., Chen, J. et al. A potentially functional polymorphism in the promoter region of miR-34b/c is associated with an increased risk for primary hepatocellular carcinoma. Int. J. Cancer 128, 412–417 (2011).
Fang, Y., Xue, J. L., Shen, Q., Chen, J. & Tian, L. MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology 55, 1852–1862 (2012).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Otsuka, M., Kishikawa, T., Yoshikawa, T. et al. MicroRNAs and liver disease. J Hum Genet 62, 75–80 (2017). https://doi.org/10.1038/jhg.2016.53
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2016.53
This article is cited by
-
MiRNA/mRNA network topology in hepatitis virus B-related liver cirrhosis reveals miR-20a-5p/340-5p as hubs initiating fibrosis
BMC Medical Genomics (2022)
-
Plasma circulating microRNAs associated with obesity, body fat distribution, and fat mass: the Rotterdam Study
International Journal of Obesity (2022)
-
microRNA profiles of serum exosomes derived from children with nonalcoholic fatty liver
Genes & Genomics (2022)
-
MicroRNA signature in hepatocellular carcinoma patients: identification of potential markers
Molecular Biology Reports (2020)
-
Suppression of miR-30a-3p Attenuates Hepatic Steatosis in Non-alcoholic Fatty Liver Disease
Biochemical Genetics (2020)