Abstract
In successive reports from 2014 to 2015, X-ray repair cross-complementing protein 4 (XRCC4) has been identified as a novel causative gene of primordial dwarfism. XRCC4 is indispensable for non-homologous end joining (NHEJ), the major pathway for repairing DNA double-strand breaks. As NHEJ is essential for V(D)J recombination during lymphocyte development, it is generally believed that abnormalities in XRCC4 cause severe combined immunodeficiency. Contrary to expectations, however, no overt immunodeficiency has been observed in patients with primordial dwarfism harboring XRCC4 mutations. Here, we describe the various XRCC4 mutations that lead to disease and discuss their impact on NHEJ and V(D)J recombination.
Similar content being viewed by others
Introduction
Cellular DNA is continuously damaged by various endogenous and exogenous factors. DNA double-strand breaks (DSBs) are the most severe form of DNA damage, which is mainly repaired by non-homologous end joining (NHEJ) or homologous recombination.1, 2 NHEJ is an inaccurate repair system that directly joins broken DSB ends with a frequent nucleotide loss at the ends, whereas homologous recombination accurately repairs DSBs by using a homologous DNA template such as a sister chromatid.1, 2 On DSB induction, NHEJ is preferentially chosen over homologous recombination in mammalian cells;1, 2 thus, cells deficient in NHEJ have reduced capacity for repairing DSBs and display enhanced radiosensitivity.3 In the NHEJ reaction, Ku protein (a heterodimer of Ku70/Ku80) binds to the ends of a DSB, followed by the recruitment of DNA-dependent protein kinase catalytic subunit and Artemis, as well as DNA polymerase λ/μ, which trim the ends. Subsequently, the ends are ligated by DNA ligase IV (LIG4) to complete the repair (Figure 1a). LIG4 is absolutely required for DSB repair via NHEJ, and other DNA ligases cannot substitute for this function.3 During lymphocyte development, NHEJ is essential for V(D)J recombination to reseal DSBs (that is, coding and signal ends) that have been produced by the action of the RAG1/RAG2 complex (Figure 1b). The X-ray repair cross-complementing protein 4 (XRCC4) protein forms a tight complex with LIG4 and contributes to LIG4 stabilization.4 XRCC4 also binds to XLF (XRCC4-like factor; also known as Cernunnos or NHEJ1), and this complex forms filaments at DSB sites to bridge the broken ends and promote efficient ligation catalyzed by LIG4 (Figure 1a).5, 6 Human XRCC4 protein consists of 336 amino acids and has three constituent domains (Figure 2a). The N-terminal head domain is involved in DNA binding and XRCC4/XLF filament formation, and the coiled-coil domain is involved in stable complex formation with LIG4.7 Details regarding the role of the C-terminal domain are unknown, although this region is suggested to be involved in intracellular localization8, 9 and in the suppression of NHEJ in the M-phase.10, 11
Non-homologous end joining (NHEJ) is important for DNA double-strand break (DSB) repair and V(D)J recombination. (a) General steps of NHEJ. The NHEJ reaction is initiated by the binding of Ku protein (a heterodimer of Ku70/Ku80) to the ends of a DSB. DNA-dependent protein kinase catalytic subunit (DNA-PKcs), Artemis and several additional factors are recruited to the Ku-DNA complex to process the ends. Finally, the ends are ligated by DNA ligase IV (LIG4), which is stabilized by and forms a complex with XRCC4. During this reaction, XRCC4/XLF filaments are formed at both sides of the DSB to bridge the two broken ends and promote efficient ligation. (b) NHEJ is essential for V(D)J recombination. V(D)J recombination is initiated by the lymphoid-specific proteins RAG1 and RAG2. The RAG1/RAG2 complex cleaves the DNA between the V, D and J segments and their flanking recombination signal sequences (RSSs) to produce two hairpinned coding ends and two signal ends. NHEJ reseals these DNA ends to complete coding and signal joint formation. A full color version of this figure is available at the Journal of Human Genetics journal online.
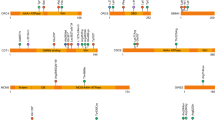
XRCC4 mutations identified in patients with primordial dwarfism. (a) Structure of human XRCC4 and mutations identified in patients with primordial dwarfism. The human XRCC4 gene consists of eight exons and is located on the long arm of chromosome 5 (5q14.2). Human XRCC4 consists of 336 amino acids and possesses three domains. Mutations predicted not to abrogate complex formation with LIG4 are shown in bold. The numbers in the protein sequence indicate exons coding for the corresponding amino acids. (b) Impact of XRCC4 mutations on LIG4-binding capacity and mRNA levels.
XRCC4 and LIG4 are both indispensable for NHEJ and appear to have similar physiological roles. Indeed, no differences have been observed between the knockout mice phenotypes (Table 1); specifically, a large amount of neuronal apoptosis is induced during late gestation in Xrcc4- and Lig4-knockout mice, leading to embryonic lethality.12, 13 These phenotypes are very different from those of mice deficient in other NHEJ factors (DNA-dependent protein kinase catalytic subunit, Artemis or XLF), which are all viable (Table 1).14 In humans, mutations of NHEJ factors have been shown to cause severe combined immunodeficiency (SCID) and microcephaly.15 For example, SCID and microcephaly occur concurrently in patients with LIG4 syndrome, which is caused by mutations in LIG4.16 Patients harboring mutations in PRKDC (which encodes DNA-dependent protein kinase catalytic subunit) or in DCLRE1C (which encodes Artemis) also exhibit SCID;16 however, microcephaly is only observed concurrently in patients possessing mutations in PRKDC,16 but not in patients with SCID caused by DCLRE1C mutations.17, 18 Mutations in XLF cause microcephaly and developmental delays similar to that observed in patients with LIG4 syndrome, although the impact of XLF deficiency on the immune system are relatively mild, and thus patients with XLF mutations present with milder symptoms than SCID.15 Unlike those patients deficient in an NHEJ factor (summarized in Table 1), individuals harboring XRCC4 mutations have not been reported until recently. However, there has been no disagreement with the recognition that abnormalities in human XRCC4 would result in similar or identical phenotypes to LIG4 syndrome patients, as was the case in the mouse.
Microcephalic primordial dwarfism caused by abnormalities in XRCC4
In 2014, XRCC4 was identified as a causative gene of primordial dwarfism by whole-exome sequencing analysis.19 Subsequently, similar reports from multiple groups have implicated XRCC4 in this disorder,20, 21, 22, 23, 24 as summarized in Tables 2 and 3. Microcephaly and dwarfism, as well as nervous system abnormalities involving higher brain dysfunction, have been found in all patients with mutations in XRCC4, and most of the patients presented with microcephalic primordial dwarfism, which is defined by the presence of both prenatal and postnatal growth restriction resulting in extreme reduction in both head circumference and height. Developmental delay, gonadal abnormalities and facial dysmorphism are also commonly found in most of the patients with mutated XRCC4 alleles (Table 2). However, none of these patients displayed any overt immunodeficiency.
XRCC4 mutations and their impact on LIG4 stability and NHEJ
As shown in Table 3, c.673C>T is one of the commonly identified mutant alleles found in the patients. As the resultant mutant XRCC4 protein (p.R225*) retains the coiled-coil domain, this mutation is predicted not to interfere with the complex formation with LIG4 (Figures 2a and b). In fact, in western blot analysis performed using patient-derived cells homozygous for this mutation (cases P1 and P2), the levels of LIG4 expression were only reduced to ~40% of that of control cells even though the mutant XRCC4 protein was undetectable.20 However, the patient-derived fibroblasts showed a prolonged retention of γ-H2AX foci after exposure to ionizing radiation. Therefore, the homozygous c.673C>T mutation is considered to result in a reduced DSB repair capacity (Table 3).
A case with the heterozygous p.R225* mutation and a c.481C>T mutation (p.R161*) on the other allele has also been reported (case P14).23 As the XRCC4 protein possessing the p.R161* mutation is considered incapable of forming complexes with LIG4 (Figure 2), the level of LIG4 expression may be decreased to a greater extent than that observed when both alleles possess the c.673C>T mutation (Table 3). Similarly, in four cases with a heterozygous c.823C>T mutation (p.R275*; cases P8–P11), LIG4 expression is predicted to be significantly reduced, as the other allele harbors a c.25delC mutation and only produces a nonfunctional protein (p.H9Tfs*8; Figure 2).
A case with homozygous c.127T>C mutations (p.W43R) has also been reported by the same group (case P13; Table 3).19, 23 The Trp43 residue is involved in the formation of the hydrophobic core of the head domain, together with the Leu36, Asp38, Ser41, Phe88, Lys90, Leu113 and Lys115 residues. Therefore, the entire protein is predicted to be unstable owing to the p.W43R mutation.23 Indeed, expression levels of XRCC4 and LIG4 were strongly reduced in patient (P13)-derived cells. In addition, the head domain of XRCC4 is important for interaction with XLF (Figure 2a), and a reduced expression of XLF was observed in this patient. Whether the p.W43R alteration affects XRCC4/XLF filament formation has not been investigated; however, analysis of the post-ionizing radiation survival rates and the number of γ-H2AX foci indicates that NHEJ-mediated DSB repair efficiency was decreased in the patient homozygous for the c.127T>C mutation (Table 3).
Several cases with changes in splicing patterns owing to single nucleotide substitutions have been reported. From alleles possessing a c.246T>G mutation, a truncated XRCC4 protein (p.[D82E;V83_S105del]) is translated (Figure 3a; cases P3 and P4).21 In reverse transcription polymerase chain reaction analysis performed using samples from the patient homozygous for this mutation (case P3), a normal size messenger RNA (mRNA) corresponding to p.D82E was not detected; instead, the main transcriptional product was an mRNA possessing the c.247_315del mutation, which is generated by splicing via a newly produced splice donor sequence. Although the impact of this mutation on XRCC4/LIG4 expression was not investigated, the plasmid re-ligation efficiency was shown to be decreased compared to that in control cells.21 It is therefore speculated that NHEJ efficiency is reduced in patients homozygous for the c.246T>G mutation (Table 3).
Aberrant transcripts associated with X-ray repair cross-complementing protein 4 (XRCC4) mutations. Shown are the impact of c.246T>G mutation (a), c.482G>A mutation (b) and c.-10-1G>T mutation (c) on XRCC4 gene expression. See text for details. A full color version of this figure is available at the Journal of Human Genetics journal online.
From alleles possessing the c.482G>A mutation, three types of transcriptional products are generated (cases P5–P7).22 In addition to p.R161Q, in which the Arg161 residue is substituted with glutamine, two types of truncated XRCC4 (p.F106Ifs*1 and p.V47Dfs*5) may be translated from alleles harboring this mutation (Figure 3b), although the expression levels of these truncated proteins have not been analyzed. However, when fused to a FLAG tag and expressed in HEK293 cells, these truncated XRCC4 proteins were not detected by an anti-FLAG antibody. It thus appears that only the p.R161Q protein is expressed in patient (P5)-derived cells with the c.482G>A mutation. This mutant protein is considered capable of forming complexes with LIG4 (Figure 2b); however, its impact on NHEJ activity has not been investigated.
A mutation of a splice acceptor (SA) site has also been reported (P12).23 As shown in Figure 3c, the c.-10-1G>T mutation disrupts the SA site of intron 1. Indeed, reverse transcription polymerase chain reaction analysis performed using patient-derived cells suggested that at least two types of mRNAs are produced that do not contain the sequence corresponding to exon 2. Clearly, these mRNAs do not code for a functional protein, as the c.-10-1G>T mutation results in a large N-terminal deletion along with a frameshift mutation downstream (p.[M1_D57del;D58Mfs*14] or p.[M1_N141del;E142Mfs*12]). In western blot analysis, however, a protein of normal size similar to wild-type XRCC4 is detected, albeit in a small quantity, suggesting that an mRNA containing exon 2 is actually generated. Although the mechanism for this mRNA generation is not clear, the AG dinucleotide present eight nucleotides downstream of the original SA site may substitutionally function as an SA site (Figure 3c).
More recently, Guo et al.24 reported the case of a patient heterozygous for c.673C>T and c.760delG mutations (case P15; note that this patient had been diagnosed with Cockayne’s syndrome). Real-time PCR analysis using patient-derived cells suggested a negligible expression from the c.673C>T allele. Along with the analysis of patient (P1/P2)-derived cells described earlier, it is likely that the majority of the mRNA possessing the c.673C>T mutation undergoes nonsense-mediated mRNA decay. Thus, an XRCC4 protein partially lacking the C-terminal domain should be translated from the c.760delG allele. This mutant protein (p.D254Mfs*68) was not detected in western blot analysis; however, LIG4 appears to be expressed in the patient cells, although the expression level is reduced to ~5–15% of control cells (Table 2), indicating that the XRCC4/LIG4 complex does exist in the patient. Guo et al.24 further performed a detailed analysis of the post-ionizing radiation survival rate and the number of γ-H2AX foci using the patient-derived cells, and reached the conclusion that the efficiency of NHEJ-mediated DSB repair is reduced (Table 3). Nevertheless, absolutely no decrease was seen in V(D)J recombination assays using plasmid substrates.24 This observation is consistent with the fact that no overt immunodeficiency has been observed in all patients harboring mutations in XRCC4, a rationale that physiological V(D)J recombination has occurred normally in the absence of normal XRCC4.
Why are patients with XRCC4 mutations immunoproficient?
As stated above, all patients possessing biallelic XRCC4 mutations show microcephaly, dwarfism and nervous system abnormalities involving higher brain dysfunction, but do not exhibit immunodeficiency. It could be argued that these XRCC4 mutations are not null, and thus a mutant (or normal) form of XRCC4 capable of binding to LIG4 is expressed in all 15 cases (Table 3). This implies that the expression of LIG4 in patient cells is not completely lost. It is important to note, however, LIG4 mutations found in LIG4 syndrome patients are also not null mutations (see below).15 In most cases with XRCC4 mutations, a reduction in DSB repair efficiency is observed, suggesting that NHEJ activity is appreciably decreased. Why, then, does V(D)J recombination occur normally in human cells lacking normal XRCC4?
Unlike DSBs induced by ionizing radiation and other genotoxic agents, DSBs induced by the RAG1/RAG2 complex are programmed to be re-joined via NHEJ during V(D)J recombination. Thus, even if filament formation by XRCC4/XLF complexes is incomplete, the two ends are protected by the RAG1/RAG2 complex, allowing LIG4 to complete the ligation reaction.25, 26 In this regard, differences in the amount of cellular DSBs may affect phenotypes (Figure 4). Specifically, DSBs induced during V(D)J recombination are limited, and therefore a sufficient amount of LIG4 is available for completing joining reactions, even when LIG4 stability is reduced (due to a decrease in XRCC4 levels). In contrast, the amount of DSBs produced in the neuronal cell differentiation process is overwhelmingly larger than that produced in V(D)J recombination. In addition, a factor equivalent to RAG1/RAG2 does not exist, and the repair of these DSBs requires not only the formation of XRCC4/XLF filaments but also a higher amount of LIG4. Hence, in neuronal cells, XRCC4 mutations interfere with DSB repair, ultimately leading to the development of microcephaly and dwarfism because of the great extent of neuronal cell death. Alternatively, or additionally, the absence of XRCC4 may be functionally compensated for by other NHEJ factors in V(D)J recombination. The recently discovered PAXX (paralog of XRCC4 and XLF) has been suggested to directly interact with Ku and contribute to the promotion of NHEJ reactions.27, 28 PAXX possesses a structure similar to XRCC4 and XLF, and the function of PAXX may compensate for the reduced NHEJ efficiency in V(D)J recombination.
Outcome of pathogenic X-ray repair cross-complementing protein 4 (XRCC4) mutations. Pathogenic XRCC4 mutations result in reduced cellular levels of XRCC4 and LIG4 proteins, leading to reduced NHEJ activity and DSB repair capacity. Nevertheless, physiological V(D)J recombination is not or little affected, while repair of DSBs in neuronal cells is considered compromised. See text for details. A full color version of this figure is available at the Journal of Human Genetics journal online.
Abnormalities in LIG4 cause LIG4 syndrome, which exhibits microcephaly and SCID.15 Similar to XRCC4, however, LIG4 has recently been reported to be a causative gene of microcephalic primordial dwarfism without immunodeficiency (note that this fact is not reflected in Table 1).29 In addition, cancer patients harboring a LIG4 mutation that impairs its adenylation and ligation activities do not exhibit SCID.30, 31 Interestingly, LIG4 syndrome patients express LIG4 proteins harboring mutations in the catalytic domain, whereas non-SCID microcephalic primordial dwarfism patients express catalytically normal LIG4 proteins with decreased complex formation with XRCC4.29 Hence, it may be that the level of cellular LIG4 activity is the determinant of SCID susceptibility when the XRCC4 or LIG4 gene is mutated.
Concluding remarks
On the basis of earlier studies using knockout mice and the biochemical analysis, XRCC4 was considered to be a stabilization factor for LIG4, and there has been no disagreement with the recognition of ‘XRCC4=LIG4’ in terms of loss-of-function phenotypes. However, unlike that observed in XRCC4-knockout mice and patients with LIG4 syndrome, it is now clear that patients with dwarfism harboring XRCC4 mutations do not develop SCID, even though all these patients do express extremely low (or undetectable) levels of mutated, yet LIG4-bindable, XRCC4 protein (Table 3). It has been reported that XRCC4 has the fastest evolutionary rate among NHEJ factors (that is, many mutations accumulate during the evolutionary process).32 Intriguingly, these mutations are concentrated in the C-terminal domain of XRCC4, and many residues in this region undergo post-translational modifications.8, 9, 10, 11 It is therefore possible that human XRCC4 may have acquired additional functions at its C-terminal domain. We expect that expression of pathogenic XRCC4 genes in XRCC4-deficient human cells or mice not only reveals the impact of XRCC4 mutations on LIG4 stability and NHEJ capacity, but will also clarify details regarding its connection to V(D)J recombination and illnesses. Therefore, the various cases referred to in this paper will provide new clues regarding the physiological functions of human XRCC4.
References
Lieber, M. R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79, 181–211 (2010).
Kakarougkas, A. & Jeggo, P. A. DNA DSB repair pathway choice: an orchestrated handover mechanism. Br. J. Radiol. 87, 20130685 (2014).
Adachi, N., Ishino, T., Ishii, Y., Takeda, S. & Koyama, H. DNA ligase IV-deficient cells are more resistant to ionizing radiation in the absence of Ku70: implications for DNA double-strand break repair. Proc. Natl Acad. Sci. USA 98, 12109–12113 (2001).
Bryans, M., Valenzano, M. C. & Stamato, T. D. Absence of DNA ligase IV protein in XR-1 cells: evidence for stabilization by XRCC4. Mutat. Res. 433, 53–58 (1999).
Mahaney, B. L., Hammel, M., Meek, K., Tainer, J. A. & Lees-Miller, S. P. XRCC4 and XLF form long helical protein filaments suitable for DNA end protection and alignment to facilitate DNA double strand break repair. Biochem. Cell Biol. 91, 31–41 (2013).
Reid, D. A., Keegan, S., Leo-Macias, A., Watanabe, G., Strande, N. T., Chang, H. H. et al. Organization and dynamics of the nonhomologous end-joining machinery during DNA double-strand break repair. Proc. Natl Acad. Sci. USA 112, E2575–E2584 (2015).
Junop, M. S., Modesti, M., Guarne, A., Ghirlando, R., Gellert, M. & Yang, W. Crystal structure of the Xrcc4 DNA repair protein and implications for end joining. EMBO J. 19, 5962–5970 (2000).
Fukuchi, M., Wanotayan, R., Liu, S., Imamichi, S., Sharma, M. K. & Matsumoto, Y. Lysine 271 but not lysine 210 of XRCC4 is required for the nuclear localization of XRCC4 and DNA ligase IV. Biochem. Biophys. Res. Commun. 461, 687–694 (2015).
Francis, D. B., Kozlov, M., Chavez, J., Chu, J., Malu, S., Hanna, M. et al. DNA Ligase IV regulates XRCC4 nuclear localization. DNA Repair 21, 36–42 (2014).
Terasawa, M., Shinohara, A. & Shinohara, M. Canonical non-homologous end joining in mitosis induces genome instability and is suppressed by M-phase-specific phosphorylation of XRCC4. PLoS Genet. 10, e1004563 (2014).
Lees-Miller, S. P. DNA double strand break repair in mitosis is suppressed by phosphorylation of XRCC4. PLoS Genet. 10, e1004598 (2014).
Gao, Y., Sun, Y., Frank, K. M., Dikkes, P., Fujiwara, Y., Seidl, K. J. et al. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell 95, 891–902 (1998).
Frank, K. M., Sekiguchi, J. M., Seidl, K. J., Swat, W., Rathbun, G. A., Cheng, H. L. et al. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature 396, 173–177 (1998).
Bunting, S. F. & Nussenzweig, A. End-joining, translocations and cancer. Nat. Rev. Cancer 13, 443–454 (2013).
Woodbine, L., Gennery, A. R. & Jeggo, P. A. The clinical impact of deficiency in DNA non-homologous end-joining. DNA Repair 16, 84–96 (2014).
Woodbine, L., Neal, J. A., Sasi, N. K., Shimada, M., Deem, K., Coleman, H. et al. PRKDC mutations in a SCID patient with profound neurological abnormalities. J. Clin. Invest. 123, 2969–2980 (2013).
Moshous, D., Callebaut, I., de Chasseval, R., Corneo, B., Cavazzana-Calvo, M., Le Deist, F. et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell 105, 177–186 (2001).
Kurosawa, A. & Adachi, N. Functions and regulation of Artemis: a goddess in the maintenance of genome integrity. J. Radiat. Res. 51, 503–509 (2010).
Shaheen, R., Faqeih, E., Ansari, S., Abdel-Salam, G., Al-Hassnan, Z. N., Al-Shidi, T. et al. Genomic analysis of primordial dwarfism reveals novel disease genes. Genome Res. 24, 291–299 (2014).
Bee, L., Nasca, A., Zanolini, A., Cendron, F., d'Adamo, P., Costa, R. et al. A nonsense mutation of human XRCC4 is associated with adult-onset progressive encephalocardiomyopathy. EMBO Mol. Med. 7, 918–929 (2015).
de Bruin, C., Mericq, V., Andrew, S. F., van Duyvenvoorde, H. A., Verkaik, N. S., Losekoot, M. et al. An XRCC4 splice mutation associated with severe short stature, gonadal failure, and early-onset metabolic syndrome. J. Clin. Endocrinol. Metab. 100, E789–E798 (2015).
Rosin, N., Elcioglu, N. H., Beleggia, F., Isguven, P., Altmuller, J., Thiele, H. et al. Mutations in XRCC4 cause primary microcephaly, short stature and increased genomic instability. Hum. Mol. Genet. 24, 3708–3717 (2015).
Murray, J. E., van der Burg, M., IJspeert, H., Carroll, P., Wu, Q., Ochi, T. et al. Mutations in the NHEJ component XRCC4 cause primordial dwarfism. Am. J. Hum. Genet. 96, 412–424 (2015).
Guo, C., Nakazawa, Y., Woodbine, L., Bjorkman, A., Shimada, M., Fawcett, H. et al. XRCC4 deficiency in human subjects causes a marked neurological phenotype but no overt immunodeficiency. J. Allergy Clin. Immunol. 136, 1007–1017 (2015).
de Villartay, J. P. When natural mutants do not fit our expectations: the intriguing case of patients with XRCC4 mutations revealed by whole-exome sequencing. EMBO Mol. Med. 7, 862–864 (2015).
Vera, G., Rivera-Munoz, P., Abramowski, V., Malivert, L., Lim, A., Bole-Feysot, C. et al. Cernunnos deficiency reduces thymocyte life span and alters the T cell repertoire in mice and humans. Mol. Cell. Biol. 33, 701–711 (2013).
Xing, M., Yang, M., Huo, W., Feng, F., Wei, L., Jiang, W. et al. Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway. Nat. Commun. 6, 6233 (2015).
Ochi, T., Blackford, A. N., Coates, J., Jhujh, S., Mehmood, S., Tamura, N. et al. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science 347, 185–188 (2015).
Murray, J. E., Bicknell, L. S., Yigit, G., Duker, A. L., van Kogelenberg, M., Haghayegh, S. et al. Extreme growth failure is a common presentation of ligase IV deficiency. Hum. Mutat. 35, 76–85 (2014).
Riballo, E., Doherty, A. J., Dai, Y., Stiff, T., Oettinger, M. A., Jeggo, P. A. et al. Cellular and biochemical impact of a mutation in DNA ligase IV conferring clinical radiosensitivity. J. Biol. Chem. 276, 31124–31132 (2001).
Riballo, E., Critchlow, S. E., Teo, S. H., Doherty, A. J., Priestley, A., Broughton, B. et al. Identification of a defect in DNA ligase IV in a radiosensitive leukaemia patient. Curr. Biol. 9, 699–702 (1999).
Demogines, A., East, A. M., Lee, J. H., Grossman, S. R., Sabeti, P. C., Paull, T. T. et al. Ancient and recent adaptive evolution of primate non-homologous end joining genes. PLoS Genet. 6, e1001169 (2010).
Acknowledgements
We thank Haruka Watabe for the discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Saito, S., Kurosawa, A. & Adachi, N. Mutations in XRCC4 cause primordial dwarfism without causing immunodeficiency. J Hum Genet 61, 679–685 (2016). https://doi.org/10.1038/jhg.2016.46
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2016.46