Abstract
As the methodology of genetic detection has developed rapidly in recent years, through techniques such as genome-wide association studies (GWAS) and the secondary generation of sequencing, we are able to view the genomic landscape more clearly. It is well known that genes have a vital role in the pathogenesis of immune-mediated diseases (IMDs), which could provide important insight into new clinical therapeutic targets. Here, we review the genomic landscape of IMDs and analyse overlapping loci between diseases. There may be a need for more epigenetics studies to aid in the understanding of the transition from genotype to phenotype.
Similar content being viewed by others
Introduction
Immune-mediated diseases (IMDs) are a clinically heterogeneous group of immune system diseases.1 Diseases such as psoriatic arthritis (PsA), coeliac disease (CeD), rheumatoid arthritis (RA), psoriasis (Ps), primary biliary cirrhosis (PBC), spondyloarthropathies (SpA), juvenile idiopathic arthritis (JIA), type 1 diabetes (T1D), autoimmune thyroid disease (AITD) and inflammatory bowel diseases (IBDs) have very different clinical presentations and appear to be unrelated, but they have been recognized as sharing common pathogenic mechanisms.2, 3, 4, 5, 6 Most IMDs are epidemic in highly developed industrialized countries and rank in the top 10 causes of death in women under 65 years of age in the United States of America.7 The National Institutes of Health has estimated that over 23.5 million Americans are afflicted with IMDs, which affect ~5–7% of Western populations.2, 8, 9 As the identification of IMDs increases, so does the expected overall prevalence increase.10
Knowledge of the pathogenic mechanisms of IMDs remains limited, and they are believed to occur as a consequence of an imbalance in the complex interplay between genetic and environmental factors.11 The marked emergence of IMDs in highly developed industrialized countries in the latter half of the 20th century suggests that environmental factors are involved in the onset of IMDs.7 Many environmental determinants of some IMDs, such as cigarette smoking, appendectomy, urbanization, pollution, diet, antibiotic use, hygiene status, socioeconomic status and microbial exposure, have been identified, but whether the relationship between those factors and IMDs is causal remains to be established.7, 12, 13, 14
IMDs are considered to share a substantial portion of their heritable aetiology.15 First, epidemiologic observations have indicated that IMDs can co-occur in either the same individual or in closely related family members, and second, clinical data have shown that the same therapies exhibit similar efficacies across diseases.3, 16, 17, 18, 19 The best known and undeniable hallmark of IMDs is their association with shared HLA haplotypes; however, variation at this locus cannot explain all of the familial risk.20 Over the past decade, significant progress has been made in the identification of genetic factors in IMDs, with associations found at convincing levels of statistical significance between over 200 common or rare variants.20, 21 These discoveries have been driven largely by genome-wide association studies (GWAS), which provide hypothesis-free surveys of the human genome for common variants associated with disease susceptibility.22, 23 The number of variants found to be associated with each disease from the GWAS data is increasing every day; however, these allelic variants together account for a relatively small risk, explaining between 20 and 50% of heritability.24, 25 The introduction of the Immunochip, which is a single-nucleotide polymorphism (SNP) microarray designed for deep replication and fine mapping, represents a step forward because it allows for the identification of genes more strictly related to the immune/inflammatory pathways.26 Many studies on IMD genetics using the Immunochip have successfully identified a significant number of common genetic variants associated with multiple IMDs and have highlighted several phenotype-specific SNPs.25 Sharing genetic loci across IMDs is not only common but also complex.
In this review, we provide an overview of advances in the genetics of various IMDs, focusing on the overlapping genetic factors shared among multiple IMDs and some disease-specific genes that exert relatively large effects. Comparing and contrasting the overlapping genetic factors between GWAS and the Immunochip studies allowed us to gain insight into the underlying biological mechanisms of these genes associated with pathogenesis of IMDs. Studying disease-specific genetics also allowed us to better understand the notion that genotype determines phenotype. We also review the ethnic differences in genetic factors and discuss what the next steps should be for the immune genetics field to leverage these observations into a more complete understanding of disease mechanisms.
Overview of genomic landscape of IMDs
Familial clustering of multiple diseases, epidemiological co-occurrence and the similar efficacy of therapies across diseases suggest that genetic factors predispose individuals to IMDs.17, 18, 27, 28 Studies have compared the concordance rates of IMDs in monozygotic twins and dizygotic twins and showed that the disease concordance was much higher in monozygotic twins compared with that in dizygotic twins.29, 30, 31 In addition, the recurrence risk for a patient’s sibling is larger compared with that for a sibling of an individual from a healthy population.27 These results indicate that genetic factors are important in the development of IMDs.
The major histocompatibility complex (MHC) loci were the first reported genetic loci associated with IMDs and remain the most strongly associated genes for most IMDs.25 MHC is an extremely gene-dense region with hundreds of immunologically active genes.27 More than 28% of the expressed transcripts from the MHC region are believed to have essential functions in the human immune system, and the strongest association with IMDs maps to the HLA region.27, 32 However, variations in the MHC loci cannot entirely explain familial risk, and other genetic determinants must also be shared in complex patterns of pathogenesis.20
Before the introduction of GWAS (pre-2006), the identification of disease-predisposing genes was based on candidate gene approaches and family-based linkage studies.27 Although there were some successful studies that clarified the role of the CTLA4, PTPN22 and NOD2 genes in IMDs, in the most cases, they were unsuccessful because of limited knowledge of disease processes and a lack of common genetic variation mapping.
After the completion of the Human Genome Project and International HapMap Project and the introduction of array-based approaches, GWAS allowed for the testing of every gene in the human genome for association with a disease or trait of interest.27, 33, 34 Hundreds of loci harbouring risk alleles for a variety of IMDs have been identified by GWAS, and one of the most striking observations from the GWAS results is the overlap in signals between different diseases.27, 35 There are also some disease-specific genetic loci that often exert relatively large effects.25 Although the number of variants found to be associated with IMDs is increasing daily, they can explain <50% of the genetic variation for each IMD. It is likely that the GWAS have missed many rare or population-specific variants that contribute to heritability.25
The Immunochip array is expected to reveal even more of the shared genetic factors because it allows for identification of genes that are more strictly related to the immune/inflammatory pathways.24, 27 With Immunochip, pathways shared by IMDs and specific genetic variants associated with different clinical features are beginning to emerge.25 Immunochip array has been used in CeD, IBD, PBC, Ps, systemic sclerosis, Takayasu artetiris and RA investigations, and it has already revealed several interesting observations.36, 37, 38, 39, 40, 41, 42
Functional data analysis, such as gene expression data and protein–protein interaction data, could incorporate functional data from GWAS loci and help to identify the potentially pathogenic cell types associated with IMDs.43, 44, 45, 46, 47 Genetically engineered mouse models could imitate phenotypes similar to IMDs and help to prioritize candidate genes in GWAS loci.27 Isis Ricañ o-Ponce and Cisca Wijmenga extracted all the lead SNPs from studies performed in Caucasian individuals and their perfect proxies and successfully annotated SNPs associated with regulatory sequences as follows: 7.6% of the variants mapped to promoter histone marks, 18.8% mapped to enhancer histone marks, 32.1% were present at DNase-hypersensitive sites, 42.3% changed motifs and 14.4% mapped to protein-bound regions.
Overlapping loci between diseases
The notion that IMDs share some of their genetic background is now well accepted, and the most striking observation from GWAS is the overlap in signals between different diseases.16, 25, 27, 48 The GWAS on 12 IMDs revealed that 68 non-HLA loci are shared across IMDs and that 11 SNPs are shared by more than four diseases.25 The introduction of the Immunochip array is expected to reveal even more of the shared genetic factors, and it allows for the identification of genes more strictly related to the immune/inflammatory pathways.24, 25, 27 A good example is provided by Van Sommeren et al.,49 who assessed shared disease pathways by performing an extensive pathway analysis of protein–protein interactions and cotranscriptional analysis and identified 370 protein–protein interactions that are shown to cluster in specific biological pathways.
The sharing of genetic loci across IMDs is not only common but also complex. For example, a shared SNP can confer increased risk for more than one disease (‘correlated and concordant’); a risk allele may appear to impart risk for some diseases but is protective against other diseases (‘correlated but opposite’); or different haplotypes may be implicated in a shared locus (‘non-correlated’).25 Here, we summarize the overlapping genetic loci and pathways that are involved in IMDs.
Major histocompatibility complex
The MHC is an extremely gene-dense region with long-range linkage disequilibrium and hundreds of immunologically active genes, including the HLA genes.25 The association of individual diseases with multiple MHC genes has long been suspected, and more than 28% of the expressed transcripts from the MHC region are believed to have essential functions in the human immune system.27, 32 Not surprisingly, the strongest association with IMDs maps to the HLA region.27 A good example is that the HLA-B27 genotype exists in 90% of AS patients and in only 2–8% of healthy Europeans.50
Some of the HLA alleles associated with IMDs are present in several IMDs. For example, seropositive diseases are typically associated with HLA class II alleles as follows: CeD, T1D, AITD and systemic lupus erythematosus (SLE) are associated with the HLA-DR3-DQ2 haplotype; and RA is associated with different alleles of HLA-DRB1.51, 52 Extended haplotypes and strong linkage disequilibrium make it difficult to identify independent association signals within the HLA haplotype and its region.27
Although classic HLA alleles constitute most of the associations across IMDs, the non-MHC genes in the MHC region are also likely to contribute to IMD heritability.25 Yang et al.53 analysed epidemiologic studies on C4A or C4B deficiencies in human SLE and found that C4A deficiencies were present in 40–60% of SLE patients from almost all ethnic groups and races investigated. The co-occurrence of IMDs is because of the influence of shared HLA haplotypes, but variation at this locus cannot entirely explain familial risk, as other genetic determinants must also be shared in complex patterns of overlap.16, 20
The IL-12 family
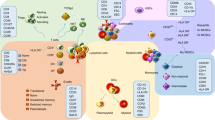
The interleukin-12 (IL-12) family comprises four heterodimeric cytokines: IL-12, IL-23, IL-27 and IL-35.11 Their respective receptors have key roles in immune responses, and a surprising number of autoimmune diseases (AIDs) have been found to be associated with genetic variation in this gene family.48 Based on the association of AIDs with the various IL-12 regions, two major clusters can be distinguished.
The first cluster shows strong associations with IL-23R, and the majority are associated with the IL-12B gene region as well.25 This result indicates an important pathogenic role for T-helper type 17 (Th17) pathway and, possibly, the T-helper type 1 (Th1) pathway. Crohn’s disease (CrD), ulcerative colitis (UC), Ps, PsA, AS and RA are all encompassed in this cluster.11 IL-23 has an important role in amplifying and stabilizing Th17 cells,54, 55 and the IL-23R gene appears to be the gene with the most consistent associations in the first cluster of AIDs. The majority of the IL-23R SNPs associated with these diseases protect against the development of these diseases, suggesting that the presence of these SNPs decreases the function of the proinflammatory Th17 cells.56 Another IL-23R variant (rs10889677) located in the 3′-untranslated region is associated with an increased disease susceptibility to IBD.56 IL-12 is produced by antigen-presenting, phagocytic and B cells in response to infection and promotes the development of naive CD4+ T cells into proinflammatory Th1 cells, which secrete the proinflammatory cytokine interferon-γ.57 The polymorphisms in the 3′-untranslated region region of the IL-12B gene include the TaqI restriction fragment length polymorphism at position 1188 (rs3212227), which is associated with multiple AIDs.58 Moreover, genetic variations in the coding region of murine IL-12b resulted in more efficient binding of the IL-12p40 variant to the IL-12p35 and IL-23p19 subunits, likely because of conformational changes resulting from differential glycosylation induced by the polymorphism.59
The second cluster is associated with polymorphisms in the IL-12A gene region, suggesting an important pathogenic role for IL-12 and/or IL-35 in the Th1/IL-35 pathway.25 PBC, CeD, multiple sclerosis (MS) and Graves’ disease are all encompassed in this cluster.11 Increased IL-12p35 mRNA levels in the small intestinal mucosa of CeD patients correlate with the rs9811792 risk allele located ~10 kb proximal to the start codon of the IL-12A gene.60 Another polymorphism (rs582054) located in an IL-12A intron is associated with atopic dermatitis and was shown to be inversely correlated with blood eosinophil count.61 However, it is still unknown whether altered IL-12A gene expression primarily affects IL-12 formation or the recently identified IL-35 expression.
In addition to the two clusters described above that categorize diseases based on their respective associations with the IL-12 gene cluster, there are additional IMDs that do not show associations strong enough to enable their categorization into either of the two clusters. Asthma, atopic dermatitis (AD) and T1D are classified in this group and may be associated with Th2.11 Th2 cells are involved in allergic disorders and protect against extracellular pathogens.62 The decreased IL-12 secretion in these AIDs results in impaired inhibition of Th2 cell differentiation, leading to an exaggerated Th2 response.63
GWAS and Immunochip studies have identified several SNPs in IL-12 family gene are associated with more than one IMD. As an example of genetic overlap between IMDs, rs11209026, a missense polymorphism in the IL-23R, is shared among CrD, UC, AS and Ps.11 Another example is that UC, CrD, AS and Ps have all been linked to an SNP located in the upstream of the 5′-untranslated region of IL-12B gene.11
Th17 pathway
Th17 cells, a distinct subset of CD4+ T cells with IL-17 as their major cytokine, orchestrate the pathogenesis of inflammatory and AIDs. Deregulated Th17 cells contribute to inflammatory and AIDs. A further understanding of the mechanistic role of Th17 cells in inflammatory and AIDs will shed light on therapeutic targets that can potentially be exploited for the management of inflammatory and AIDs. For example, the frequency of Th17 cells is higher in patients with RA compared with that in healthy controls, suggesting that a Th17/Treg (regulatory T cell) cell imbalance may contribute to the pathogenesis and progression of RA.64 In this disease, Th17 cells represent a proinflammatory subset, whereas Treg cells have an antagonistic effect. Studies have demonstrated that IL-17 upregulates the production of IL-1β and tumour necrosis factor-α (TNFα) in antigen-presenting cells in arthritic joints.65 Similarly, IL-17 and Th17 cells have a role in Behcet's disease and uveitis,66 demonstrating that Th17 cells participate in the pathogenesis of AIDs. Another AID is SLE, which is a complex disease of unknown aetiology. However, multiple links between SLE and Th17 have emerged.67 A strong correlation between Th17 cell expression and SLE disease activity has been found, with a higher percentage of CD3+CD4+ T cells producing IL-17 in SLE patients compared with healthy controls.68 In Sjögren syndrome (SS), IL-17 and IL-23 levels are increased in the salivary gland as well as in the serum of patients. These cytokines and their receptors were also shown to be expressed within lymphocytic infiltrates and ductal areas in the salivary glands of patients.69, 70, 71, 72, 73 There is evidence that IL-17 has a crucial pathogenic role; thus, it may serve as a potential therapeutic target for amelioration of Sjögren syndrome. In Ps, which is a common skin disorder, IL-17A acts on keratinocytes to induce the expression of CCL20, thereby recruiting CCR6t Th17 cells and dendritic cells to the skin74, 75 and potentially providing a positive feedback loop that results in the maintenance of these cells in psoriatic lesions.76 Experimental evidence is also available to support a role for IL-17A in other immune inflammatory diseases, including PsA, AS, IDB, CrD, MS, diabetes and asthma.77, 78, 79, 80 T1D involves CD4+ and CD8+ T-cell-mediated destruction of pancreatic β-cells, and the uncontrolled expansion of Th17 cells is involved in the T1D pathology of the disease.81 Moreover, in patients with allergic asthma, the number of Th17 cells in peripheral blood, sputum and bronchoalveolar lavage fluids is increased compared with that in healthy controls, and levels of Th17 cells positively correlate with the severity of airway remodelling.82, 83 Above all, there is accumulating evidence that Th17 cells regulate many types of inflammatory diseases and AIDs. An understanding of the role of Th17 in these conditions will provide important insights and elucidate novel targets for therapeutic intervention.
TNF and its superfamily
TNF is one of a large group of cytokines collectively named the ‘TNF superfamily’, which includes cytokines that share molecular and functional similarities. Most of these cytokines are involved in the regulation of several steps of the biological processes related to inflammatory and immune responses.84, 85
Extensive preclinical and clinical investigations have shown that TNF has a pivotal role in the pathogenesis and pathophysiology of IMDs, which has been confirmed by the efficacy of anti-TNF biotechnological drugs, such as etanercept, infliximab and adalimumab, in the therapeutic management of these disorders.2, 86 Overexpression of TNF has been shown to promote proinflammatory processes. In particular, along with the dysregulation of other cytokines and a variety of cell types, TNF is implicated in the pathogenesis of IMDs, such as RA, CrD, Ps, PsA, SLE, T1D, MS, asthma, allergy and UC.87
In preclinical and clinical RA studies, it was shown that abnormal elevations of TNF concentrations at inflammatory sites were a primary factor in disease activity, and these observations generated the hypothesis that the removal of excess TNF from inflamed joints would confer therapeutic benefits.88 To support these concepts, transgenic mice with an overexpression of TNF were found to spontaneously develop an arthritic pathology that displayed clinical and histological features similar to those of RA.89 In addition, in an experimental model of collagen-induced arthritis, the blockade of TNF was effective in inhibiting disease activity.90 In experimental and clinical studies of the pathogenesis of IBDs, it was suggested that there are elevated levels of TNF in the inflamed mucosa of patients with CrD,91 as well as that enhanced levels of TNF are present in patients with both CrD and UC.92 TNF is also involved in the mechanisms underlying the pathogenesis of both Ps and PsA.93 With regard to Ps, TNF may be involved in the disease pathogenesis, including stimulation of the maturation of Langerhans cells and dendritic cells, with skewing of lymphocyte differentiation;94 promotion of dendritic cell migration from the skin to lymph nodes;95 accumulation of leucocytes in the inflamed skin through the induction of adhesion molecules and chemokines on dermal microvascular endothelial cells, keratinocytes and dermal firoblasts;96 induction of dermal vascular changes via the production of vascular endothelial growth factor by keratinocytes and hyperproliferation of keratinocytes;97 and induction of itching through the activation of TNF receptors on sensory nerve endings.93 In PsA, it has been shown that TNF has a primary role in the induction of inflammation and joint and bone damage through the following mechanisms: production of lytic enzymes, such as matrix metalloproteases;98 contribution to synovial vascular proliferation by the induction of angiogenic growth factors; stimulation of bone resorption; inhibition of bone formation; and inhibition of the synthesis of proteoglycans, with the subsequent occurrence of bone erosions leading to osteolysis, new bone deposition or both.99
TNF is implicated in the pathogenesis of many IMDs, such as RA, CrD, Ps, PsA, SLE, T1D, MS, asthma, allergy and UC,87 whereas GWAS and Immunochip studies have revealed that the reason might be TNFα-induced protein 3 (TNFAIP3) locus. TNFAIP3, which is also known as A20 and induced rapidly by TNF, is associated with RA, CeD, T1D, SLE, scleroderma, Ps, CrD and UC.25
Opposite alleles
The phenomenon that some SNPs show the strongest association across IMDs but act in opposite manners indicates that the genetic architecture of immune-related diseases may be even more complex than previously thought.27 Several risk alleles appear to impart risk for some diseases but are protective against others.100, 101 A well-known example in the literature is the R620 W variant in PTPN22, known to confer risk for T1D102 and the development of autoantibodies,103 RA104 and vitiligo105 but to be protective against CrD.106 Another emerging example is the rs744166 polymorphism in an intron of signal transducer and activator of transcription 3 on chromosome 17, where the A allele confers risk for CrD106 and the G allele confers risk for MS.107 Treatments targeting one IMD may inadvertently provoke or exacerbate another.25 For example, the rs1800696 variant of TNF receptor 1 is protective against AS but increases the risk for MS.108
Currently, the mechanisms of these opposing risk effects remain unclear. These opposing allelic effects may be worth investigating, as they could help us better understand the underlying disease biology.27
Ethnicity
Different ethnic groups may contribute to genetic diversity for gene mapping. For example, in Caucasians, IL-23R polymorphisms have been identified in CrD and AS. However, IL-23R has not been shown to be associated with CrD in East Asians.109 In 2009, we confirmed that rs11209026, which has been identified as the key IL-23R polymorphism associated with AS, is not polymorphic in the Han Chinese population.110
In several other IMDs, the polymorphic status of SNPs has contributed to differing levels of association between ethnic groups. PTPN22 is strongly associated with RA in Caucasians; however, it has been shown that the C1858T SNP, which is thought to be the key SNP associated with the disease in European Caucasians, is not polymorphic in East Asians, explaining the lack of association of PTPN22 with RA in these populations.111 Similarly, the NOD2/CARD15 polymorphisms associated with CrD in European Caucasian populations are not polymorphic in Asians.
These findings suggest that there are significant differences in susceptibility genes between different ethnicities, which could be exploited in an effort to distinguish true disease-associated polymorphisms from linkage disequilibrium effects.
From genotype to phenotype
Although the phenomenon of overlapping loci between IMDs is very common, there are some convincingly phenotype-specific loci. For example, in contrast to seropositive diseases, which are typically associated with HLA class II alleles,51, 52 seronegative diseases, such as CrD, Ps and AS, are generally associated with HLA class I alleles, which tend to be disease-specific.25 HLA B27 is confirmed to be associated with AS; HLA Cw6 is closely related to Ps; and HLA B51 is strongly linked to Behcet's disease.112, 113, 114, 115, 116 CrD is associated with SNPs in the MHC class I region, but it is not associated with any classical HLA allele.25, 39 There are also non-HLA examples for seronegative diseases; specifically, NOD2 (nucleotide-binding oligomerization domain-containing 2) and ATG16L1 (autophagy-related 16-like 1) are associated with CrD, and HNF4A is strongly linked to UC.117, 118, 119 Furthermore, all three are confirmed to have a vital role in the pathogenesis of CrD and UC.117, 118, 119, 120, 121, 122
There are also some phenotype-specific gene loci associated with seropositive IMDs. For example, INS (encoding insulin) was found to be associated with T1D, and TSHR (encoding thyroid-stimulating hormone receptor) was closely associated with Graves’ disease.123 PRTN3 (encoding proteinase 3) was confirmed to be strongly linked to antineutrophil cytoplasmic antibody-associated vasculitis, and PADI4 (encoding peptidyl arginine deiminase, type IV) is highly associated with RA.124, 125
Another good example was derived from TNF-overexpressing mouse models. Overexpression of human TNF results in the spontaneous development of severe systemic inflammation and destructive polysynovitis in mice, and the model phenocopies human RA rather than SpA.89 However, the blockade of DKK-1, an inhibitor of the Wnt signalling pathway, reverses the destructive phenotype of this model into a remodelling phenotype characterized by new bone formation in peripheral and sacroiliac joints, suggesting that TNF overexpression may partially photocopy human SpA under specific conditions.89, 126, 127, 128
An interesting phenomenon is that the disease-specific gene loci often confer large effects. Given the phenotype specificity of these loci, their generally large effect sizes, and their links to specific pathogenic pathways, it is likely that they have key phenotype-determining roles in each IMD.25 Furthermore, targeting disease-specific pathways may yield effective, disease-specific therapies with less systemic toxicity.
Conclusion and future directions
In conclusion, GWAS and the Immunochip have facilitated the discovery of disease-associated SNPs in IMDs, but several limitations exist regarding the GWAS and Immunochip studies conducted to date.
First, almost all of the studies were performed on cohorts of Caucasian descent, although a few were performed on Ashkenazi Jews and Chinese, Japanese and Korean individuals. More studies should be designed to analyse different ethnic groups.
Second, a large number of shared risk SNPs have been found across IMDs, but only a few of the IMD loci seem to be unique to a single disease. Future research should also focus on these disease-specific factors, as they may yield important clues regarding disease mechanisms and may provide avenues for prevention and/or treatment.
Third, the studies of epigenetic interaction would be helpful for assessing the expression of single alleles in relevant tissues. The significant insights gained from genetics will yield information on the genes and pathways that are associated with each disease. However, environmental factors or epigenetic events will alter the transition process from genotype to phenotype, which will be addressed in our next study.
Finally, more studies should focus on more personalized therapeutic approaches. Okada et al.129 performed a GWAS meta-analysis in European and Asian ancestries, identifying 98 biological candidate genes as the targets of approved therapies for RA and further suggesting that drugs approved for other indications may be repurposed for the treatment of RA. This comprehensive genetic study can provide important information for drug discovery.129
References
Shurin, M. R. & Smolkin, Y. S. Immune-mediated diseases: where do we stand? Adv. Exp. Med. Biol. 601, 3–12 (2007).
Blandizzi, C., Gionchetti, P., Armuzzi, A., Caporali, R., Chimenti, S., Cimaz, R. et al. The role of tumour necrosis factor in the pathogenesis of immune-mediated diseases. Int. J. Immunopathol. Pharmacol. 27 (suppl. 1), 1–10 (2014).
Torfs, C. P., King, M. C., Huey, B., Malmgren, J. & Grumet, F. C. Genetic interrelationship between insulin-dependent diabetes mellitus, the autoimmune thyroid diseases, and rheumatoid arthritis. Am. J. Hum. Genet. 38, 170–187 (1986).
Lin, J. P., Cash, J. M., Doyle, S. Z., Peden, S., Kanik, K., Amos, C. I. et al. Familial clustering of rheumatoid arthritis with other autoimmune diseases. Hum. Genet. 103, 475–482 (1998).
Hemminki, K., Li, X., Sundquist, K. & Sundquist, J. Shared familial aggregation of susceptibility to autoimmune diseases. Arthritis Rheum. 60, 2845–2847 (2009).
Thjodleifsson, B., Geirsson, A. J., Björnsson, S. & Bjarnason, I. A common genetic background for inflammatory bowel disease and ankylosing spondylitis: a genealogic study in Iceland. Arthritis Rheum. 56, 2633–2639 (2007).
Elliott, D. E. & Weinstock, J. V. Helminth–host immunological interactions: prevention and control of immune-mediated diseases. Ann. NY Acad. Sci. 1247, 83–96 (2012).
Robinson, D. Jr, Hackett, M., Wong, J., Kimball, A. B., Cohen, R. & Bala, M. IMID Study Group Co-occurrence and comorbidities in patients with immune-mediated inflammatory disorders: an exploration using US healthcare claims data, 2001-2002. Curr. Med. Res. Opin. 22, 989–1000 (2006).
El-Gabalawy, H., Guenther, L. C. & Bernstein, C. N. Epidemiology of immune-mediated inflammatory diseases: incidence, prevalence, natural history, and comorbidities. J. Rheumatol. Suppl. 85, 2–10 (2010).
Vizcarra, C. New perspectives and emerging therapies for immune-mediated inflammatory disorders. J. Infus. Nurs. 26, 319–325 (2003).
Van Wanrooij, R. L., Zwiers, A., Kraal, G. & Bouma, G. Genetic variations in interleukin-12 related genes in immune-mediated diseases. J. Autoimmun. 39, 359–368 (2012).
Ng, S. C., Bernstein, C. N., Vatn, M. H., Lakatos, P. L., Loftus, E. V. Jr, Tysk, C. et al. Geographical variability and environmental risk factors in inflammatory bowel disease. Gut. 62, 630–649 (2013).
Versini, M., Jeandel, P. Y., Rosenthal, E. & Shoenfeld, Y. Obesity in autoimmune diseases: not a passive bystander. Autoimmun. Rev. 13, 981–1000 (2014).
Padyukov, L., Silva, C., Stolt, P., Alfredsson, L. & Klareskog, L. A gene–environment interaction between smoking and shared epitope genes in HLA-DR provides a high risk of seropositive rheumatoid arthritis. Arthritis Rheum. 50, 3085–3092 (2004).
Diaz-Gallo, L. M. & Martin, J. Common genes in autoimmune diseases: a link between immune-mediated diseases. Expert Rev. Clin. Immunol. 8, 107–109 (2012).
Voight, B. F. & Cotsapas, C. Human genetics offers an emerging picture of common pathways and mechanisms in autoimmunity. Curr. Opin. Immunol. 24, 552–557 (2012).
Criswell, L. A., Pfeiffer, K. A., Lum, R. F., Gonzales, B., Novitzke, J., Kern, M. et al. Analysis of families in the multiple autoimmune disease genetics consortium (MADGC) collection: the PTPN22 620W allele associates with multiple autoimmune phenotypes. Am. J. Hum. Genet. 76, 561–571 (2005).
Eaton, W. W., Rose, N. R., Kalaydjian, A., Pedersen, M. G. & Mortensen, P. B. Epidemiology of autoimmune diseases in Denmark. J. Autoimmun. 29, 1–9 (2007).
Becker, K. G. The common genetic hypothesis of autoimmune/inflammatory disease. Curr. Opin. Allergy Clin. Immunol. 1, 399–405 (2001).
Risch, N. Assessing the role of HLA-linked and unlinked determinants of disease. Am. J. Hum. Genet. 40, 1–14 (1987).
Altshuler, D. & Daly, M. Guilt beyond a reasonable doubt. Nat. Genet. 39, 813–815 (2007).
Wellcome Trust Case–Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007).
Wellcome Trust Case Control Consortium; Australo-Anglo-American Spondylitis Consortium (TASC), Burton, P. R., Clayton, D. G., Cardon, L. R., Craddock, N. et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat. Genet. 39, 1329–1337 (2007).
Sorrentino, R. Genetics of autoimmunity: an update. Immunol. Lett. 158, 116–119 (2014).
Parkes, M., Cortes, A., van Heel, D. A. & Brown, M. A. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat. Rev. Genet. 14, 661–673 (2013).
International Genetics of Ankylosing Spondylitis Consortium (IGAS), Cortes, A., Hadler, J., Pointon, J. P., Robinson, P. C., Karaderi, T. et al, Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat. Genet. 45, 730–738 (2013).
Ricaño-Ponce, I. & Wijmenga, C. Mapping of immune-mediated disease genes. Annu. Rev. Genom. Hum. Genet. 14, 325–353 (2013).
Hauser, S. L. Multiple lessons for multiple sclerosis. N. Engl. J. Med. 359, 1838–1841 (2008).
Kaprio, J., Tuomilehto, J., Koskenvuo, M., Romanov, K., Reunanen, A., Eriksson, J. et al. Concordance for type 1 (insulin-dependent) and type 2 (non-insulin-dependent) diabetes mellitus in a population-based cohort of twins in Finland. Diabetologia 35, 1060–1067 (1992).
Kyvik, K. O., Green, A. & Beck-Nielsen, H. Concordance rates of insulin dependent diabetes mellitus: a population based study of young Danish twins. BMJ 311, 913–917 (1995).
Ebers, G. C., Bulman, D. E., Sadovnick, A. D., Paty, D. W., Warren, S., Hader, W. et al. A population-based study of multiple sclerosis in twins. N. Engl. J. Med. 315, 1638–1642 (1986).
Horton, R., Wilming, L., Rand, V., Lovering, R. C., Bruford, E. A., Khodiyar, V. K. et al. Gene map of the extended human MHC. Nat. Rev. Genet. 5, 889–899 (2004).
International HapMap Consortium A haplotype map of the human genome. Nature 437, 1299–1320 (2005).
Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Cotsapas, C. & Hafler, D. A. Immune-mediated disease genetics: the shared basis of pathogenesis. Trends Immunol. 34, 22–26 (2013).
Eyre, S., Bowes, J., Diogo, D., Lee, A., Barton, A., Martin, P. et al. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat. Genet. 44, 1336–1340 (2012).
Tsoi, L. C., Spain, S. L., Knight, J., Ellinghaus, E., Stuart, P. E., Capon, F. et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat. Genet. 44, 1341–1348 (2012).
Liu, J. Z., Almarri, M. A., Gaffney, D. J., Mells, G. F., Jostins, L., Cordell, H. J. et al. Dense fine-mapping study identifies new susceptibility loci for primary biliary cirrhosis. Nat. Genet. 44, 1137–1141 (2012).
Jostins, L., Ripke, S., Weersma, R. K., Duerr, R. H., McGovern, D. P., Hui, K. Y. et al. Host–-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012).
Trynka, G., Hunt, K. A., Bockett, N. A., Romanos, J., Mistry, V., Szperl, A. et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat. Genet. 43, 1193–1201 (2011).
Mayes, M. D., Bossini-Castillo, L., Gorlova, O., Martin, J. E., Zhou, X., Chen, W. V. et al. Immunochip analysis identifies multiple susceptibility loci for systemic sclerosis. Am. J. Hum. Genet. 94, 47–61 (2014).
Saruhan-Direskeneli, G., Hughes, T., Aksu, K., Keser, G., Coit, P., Aydin, S. Z. et al. Identification of multiple genetic susceptibility loci in Takayasu arteritis. Am. J. Hum. Genet. 93, 298–305 (2013).
Franke, L., van Bakel, H., Fokkens, L., de Jong, E. D., Egmont-Petersen, M. & Wijmenga, C. Reconstruction of a functional human gene network, with an application for prioritizing positional candidate genes. Am. J. Hum. Genet. 78, 1011–1025 (2006).
Lee, I., Blom, U. M., Wang, P. I., Shim, J. E. & Marcotte, E. M. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res. 21, 1109–1121 (2011).
Raychaudhuri, S., Plenge, R. M., Rossin, E. J. & Ng, A. C. International Schizophrenia Consortium, Purcell, S. M. et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 5, e1000534 (2009).
Raychaudhuri, S., Thomson, B. P., Remmers, E. F., Eyre, S., Hinks, A., Guiducci, C. et al. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat. Genet. 41, 1313–1318 (2009).
Rossin, E. J., Lage, K., Raychaudhuri, S., Xavier, R. J., Tatar, D., Benita, Y. et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 7, e1001273 (2011).
Cotsapas, C., Voight, B. F., Rossin, E., Lage, K., Neale, B. M., Wallace, C. et al. Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet. 7, e1002254 (2011).
Van Sommeren, S., Janse, M., Karjalainen, J., Fehrmann, R., Franke, L., Fu, J. et al. Extraintestinal manifestations and complications in inflammatory bowel disease: from shared genetics to shared biological pathways. Inflamm. Bowel Dis. 20, 987–994 (2014).
Middleton, D., Menchaca, L., Rood, H. & Komerofsky, R. New allele frequency database: http://www.allelefrequencies.net. Tissue Antigens. 61, 403–407 (2003).
Price, P., Witt, C., Allcock, R., Sayer, D., Garlepp, M., Kok, C. C. et al. The genetic basis for the association of the 8.1 ancestral haplotype (A1, B8, DR3) with multiple immunopathological diseases. Immunol. Rev. 167, 257–274 (1999).
Gregersen, P. K., Silver, J. & Winchester, R. J. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 30, 1205–1213 (1987).
Yang, Y., Chung, E. K., Zhou, B., Lhotta, K., Hebert, L. A., Birmingham, D. J. et al. The intricate role of complement component C4 in human systemic lupus erythematosus. Curr. Dir. Autoimmun. 7, 98–132 (2004).
Capon, F., Di Meglio, P., Szaub, J., Prescott, N. J., Dunster, C., Baumber, L. et al. Sequence variants in the genes for the interleukin-23 receptor (IL23R) and its ligand (IL12B) confer protection against psoriasis. Hum. Genet. 122, 201–206 (2007).
Nair, R. P., Duffin, K. C., Helms, C., Ding, J., Stuart, P. E., Goldgar, D. et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat. Genet. 41, 199–204 (2009).
Di Meglio, P., Di Cesare, A., Laggner, U., Chu, C. C., Napolitano, L., Villanova, F. et al. The IL23R R381Q gene variant protects against immune-mediated diseases by impairing IL-23-induced Th17 effector response in humans. PLoS One 6, e17160 (2011).
Trinchieri, G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol. 3, 133–146 (2003).
Zwiers, A., Kraal, L., van de Pouw Kraan, T. C., Wurdinger, T., Bouma, G. & Kraal, G. Cutting edge: a variant of the IL-23R gene associated with inflammatory bowel disease induces loss of microRNA regulation and enhanced protein production. J. Immunol. 188, 1573–1577 (2012).
Zwiers, A., Fuss, I. J., Seegers, D., Konijn, T., Garcia-Vallejo, J. J., Samsom, J. N. et al. A polymorphism in the coding region of Il12b promotes IL-12p70 and IL-23 heterodimer formation. J. Immunol. 186, 3572–3580 (2011).
Plaza-Izurieta, L., Castellanos-Rubio, A., Irastorza, I., Fernández-Jimenez, N. & Gutierrez, G. CEGEC Revisiting genome wide association studies (GWAS) in coeliac disease: replication study in Spanish population and expression analysis of candidate genes. J. Med. Genet. 48, 493–496 (2011).
Namkung, J. H., Lee, J. E., Kim, E., Kim, S., Kim, S., Shin, E. S. et al. Association of single nucleotide polymorphisms in the IL-12 (IL-12A and B) and IL-12 receptor (IL-12Rbeta1 and beta2) genes and gene-gene interactions with atopic dermatitis in Koreans. J. Dermatol. Sci. 57, 199–206 (2010).
Gavett, S. H., O'Hearn, D. J., Li, X., Huang, S. K., Finkelman, F. D. & Wills-Karp, M. Interleukin 12 inhibits antigen-induced airway hyperresponsiveness, inflammation, and Th2 cytokine expression in mice. J. Exp. Med. 182, 1527–1536 (1995).
Hofstra, C. L., Van Ark, I., Hofman, G., Kool, M., Nijkamp, F. P. & Van Oosterhout, A. J. Prevention of Th2-like cell responses by coadministration of IL-12 and IL-18 is associated with inhibition of antigen-induced airway hyperresponsiveness, eosinophilia, and serum IgE levels. J. Immunol. 161, 5054–5060 (1998).
Niu, Q., Cai, B., Huang, Z. C., Shi, Y. Y. & Wang, L. L. Disturbed Th17/Treg balance in patients with rheumatoid arthritis. Rheumatol. Int. 32, 2731–2736 (2012).
Lubberts, E. IL-17/Th17 targeting: on the road to prevent chronic destructive arthritis? Cytokine. 41, 84–91 (2008).
Zhou, Z. Y., Chen, S. L., Shen, N. & Lu, Y. Cytokines and Behcet's disease. Autoimmun. Rev. 11, 699–704 (2012).
Fransen, J. H., van der Vlag, J., Ruben, J., Adema, G. J., Berden, J. H. & Hilbrands, L. B. The role of dendritic cells in the pathogenesis of systemic lupus erythematosus. Arthritis Res. Ther. 12, 207 (2010).
Yang, J., Chu, Y., Yang, X., Gao, D., Zhu, L., Yang, X. et al. Th17 and natural Treg cell population dynamics in systemic lupus erythematosus. Arthritis Rheum. 60, 1472–1483 (2009).
Nguyen, C. Q., Hu, M. H., Li, Y., Stewart, C. & Peck, A. B. Salivary gland tissue expression of interleukin-23 and interleukin-17 in Sjögren's syndrome: findings in humans and mice. Arthritis Rheum. 58, 734–743 (2008).
Sakai, A., Sugawara, Y., Kuroishi, T., Sasano, T. & Sugawara, S. Identification of IL-18 and Th17 cells in salivary glands of patients with Sjögren's syndrome, and amplification of IL-17-mediated secretion of inflammatory cytokines from salivary gland cells by IL-18. J. Immunol. 181, 2898–2906 (2008).
Mieliauskaite, D., Dumalakiene, I., Rugiene, R. & Mackiewicz, Z. Expression of IL-17, IL-23 and their receptors in minor salivary glands of patients with primary Sjögren's syndrome. Clin. Dev. Immunol. 2012, 187258 (2012).
Miletic, M., Stojanovic, R., Pajic, O., Bugarski, D., Mojsilovic, S., Cokic, V. et al. Serum interleukin-17 & nitric oxide levels in patients with primary Sjögren's syndrome. Indian J. Med. Res. 135, 513–519 (2012).
Katsifis, G. E., Rekka, S., Moutsopoulos, N. M., Pillemer, S. & Wahl, S. M. Systemic and local interleukin-17 and linked cytokines associated with Sjögren's syndrome immunopathogenesis. Am. J. Pathol. 175, 1167–1177 (2009).
Homey, B., Dieu-Nosjean, M. C., Wiesenborn, A., Massacrier, C., Pin, J. J., Oldham, E. et al. Up-regulation of macrophage inflammatory protein-3 alpha/CCL20 and CC chemokine receptor 6 in psoriasis. J. Immunol. 164, 6621–6632 (2000).
Nograles, K. E., Zaba, L. C., Guttman-Yassky, E., Fuentes-Duculan, J., Suárez-Fariñas, M., Cardinale, I. et al. Th17 cytokines interleukin (IL)-17 and IL-22 modulate distinct inflammatory and keratinocyte-response pathways. Br. J. Dermatol. 159, 1092–1102 (2008).
Noack, M. & Miossec, P. Th17 and regulatory T cell balance in autoimmune and inflammatory diseases. Autoimmun. Rev. 13, 668–677 (2014).
Onishi, R. M. & Gaffen, S. L. Interleukin-17 and its target genes: mechanisms of interleukin-17 function in disease. Immunology 129, 311–321 (2010).
Raychaudhuri, S. P. Role of IL-17 in psoriasis and psoriatic arthritis. Clin. Rev. Allergy Immunol. 44, 183–193 (2013).
Zepp, J., Wu, L. & Li, X. IL-17 receptor signaling and T helper 17-mediated autoimmune demyelinating disease. Trends Immunol. 32, 232–239 (2011).
Wang, Y. H. & Wills-Karp, M. The potential role of interleukin-17 in severe asthma. Curr. Allergy Asthma Rep. 11, 388–394 (2011).
Shao, S., He, F., Yang, Y., Yuan, G., Zhang, M. & Yu, X. Th17 cells in type 1 diabetes. Cell Immunol. 280, 16–21 (2012).
Molet, S., Hamid, Q., Davoine, F., Nutku, E., Taha, R., Pagé, N. et al. IL-17 is increased in asthmatic airways and induces human bronchial fibroblasts to produce cytokines. J. Allergy Clin. Immunol. 108, 430–438 (2001).
Wang, Q., Li, H., Yao, Y., Xia, D. & Zhou, J. The overexpression of heparin-binding epidermal growth factor is responsible for Th17-induced airway remodeling in an experimental asthma model. J. Immunol. 185, 834–841 (2010).
Mewar, D. & Wilson, A. G. Treatment of rheumatoid arthritis with tumour necrosis factor inhibitors. Br. J. Pharmacol. 162, 785–791 (2011).
Remouchamps, C., Boutaffala, L., Ganeff, C. & Dejardin, E. Biology and signal transduction pathways of the lymphotoxin-αβ/LTβR system. Cytokine Growth Factor Rev. 22, 301–310 (2011).
Nash, P. T. & Florin, T. H. Tumour necrosis factor inhibitors. Med. J. Aust. 183, 205–208 (2005).
Kuek, A., Hazleman, B. L. & Ostör, A. J. Immune-mediated inflammatory diseases (IMIDs) and biologic therapy: a medical revolution. Postgrad. Med. J. 83, 251–260 (2007).
Knight, D. M., Trinh, H., Le, J., Siegel, S., Shealy, D., McDonough, M. et al. Construction and initial characterization of a mouse-human chimeric anti-TNF antibody. Mol. Immunol. 30, 1443–1453 (1993).
Keffer, J., Probert, L., Cazlaris, H., Georgopoulos, S., Kaslaris, E., Kioussis, D. et al. Transgenic mice expressing human tumour necrosis factor: a predictive genetic model of arthritis. EMBO J. 10, 4025–4031 (1991).
Williams, R. O., Feldmann, M. & Maini, R. N. Anti-tumor necrosis factor ameliorates joint disease in murine collagen-induced arthritis. Proc. Natl Acad. Sci. USA 89, 9784–9788 (1992).
Papadakis, K.A. & Targan, S.R. Tumor necrosis factor: biology and therapeutic inhibitors. Gastroenterology 119, 1148–1157 (2000).
Murch, S. H., Braegger, C. P., Walker-Smith, J. A. & MacDonald, T. T. Location of tumour necrosis factor alpha by immunohistochemistry in chronic inflammatory bowel disease. Gut. 34, 1705–1709 (1993).
Fantuzzi, F., Del Giglio, M., Gisondi, P. & Girolomoni, G. Targeting tumor necrosis factor alpha in psoriasis and psoriatic arthritis. Expert Opin. Ther. Targets 12, 1085–1096 (2008).
Chomarat, P., Dantin, C., Bennett, L., Banchereau, J. & Palucka, A. K. TNF skews monocyte differentiation from macrophages to dendritic cells. J. Immunol. 171, 2262–2269 (2003).
Cumberbatch, M., Dearman, R. J., Griffiths, C. E. & Kimber, I. Langerhans cell migration. Clin. Exp. Dermatol. 25, 413–418 (2000).
Mascia, F., Mariani, V., Girolomoni, G. & Pastore, S. Blockade of the EGF receptor induces a deranged chemokine expression in keratinocytes leading to enhanced skin inflammation. Am. J. Pathol. 163, 303–312 (2003).
Pastore, S., Mascia, F., Mariani, V. & Girolomoni, G. The epidermal growth factor receptor system in skin repair and inflammation. J. Invest. Dermatol. 128, 1365–1374 (2008).
Koshy, P. J., Henderson, N., Logan, C., Life, P. F., Cawston, T. E. & Rowan, A. D. Interleukin 17 induces cartilage collagen breakdown: novel synergistic effects in combination with proinflammatory cytokines. Ann. Rheum. Dis. 61, 704–713 (2002).
O' Gradaigh, D., Ireland, D., Bord, S. & Compston, J. E. Joint erosion in rheumatoid arthritis: interactions between tumour necrosis factor alpha, interleukin 1, and receptor activator of nuclear factor kappaB ligand (RANKL) regulate osteoclasts. Ann. Rheum. Dis. 63, 354–359 (2004).
Sirota, M., Schaub, M. A., Batzoglou, S., Robinson, W. H. & Butte, A. J. Autoimmune disease classification by inverse association with SNP alleles. PLoS Genet. 5, e1000792 (2009).
Wang, K., Baldassano, R., Zhang, H., Qu, H. Q., Imielinski, M., Kugathasan, S. et al. Comparative genetic analysis of inflammatory bowel disease and type 1 diabetes implicates multiple loci with opposite effects. Hum. Mol. Genet. 19, 2059–2067 (2010).
Bottini, N., Musumeci, L., Alonso, A., Rahmouni, S., Nika, K., Rostamkhani, M. et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat. Genet. 36, 337–338 (2004).
Plagnol, V., Howson, J. M., Smyth, D. J., Walker, N., Hafler, J. P., Wallace, C. et al. Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLoS Genet. 7, e1002216 (2011).
Begovich, A. B., Carlton, V.E., Honigberg, L. A., Schrodi, S. J., Chokkalingam, A. P., Alexander, H. C. et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am. J. Hum. Genet. 75, 330–337 (2004).
Jin, Y., Birlea, S. A., Fain, P. R., Gowan, K., Riccardi, S. L., Holland, P. J. et al. Variant of TYR and autoimmunity susceptibility loci in generalized vitiligo. N. Engl. J. Med. 362, 1686–1697 (2010).
Barrett, J. C., Hansoul, S., Nicolae, D. L., Cho, J. H., Duerr, R. H., Rioux, J. D. et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat. Genet. 40, 955–962 (2008).
Jakkula, E., Leppä, V., Sulonen, A. M., Varilo, T., Kallio, S., Kemppinen, A. et al. Genome-wide association study in a high-risk isolate for multiple sclerosis reveals associated variants in STAT3 gene. Am. J. Hum. Genet. 86, 285–291 (2010).
Gregory, A. P., Dendrou, C. A., Attfield, K. E., Haghikia, A., Xifara, D. K., Butter, F. et al. TNF receptor 1 genetic risk mirrors outcome of anti-TNF therapy in multiple sclerosis. Nature 488, 508–511 (2012).
Yamazaki, K., Onouchi, Y., Takazoe, M., Kubo, M., Nakamura, Y. & Hata, A. Association analysis of genetic variants in IL23R, ATG16L1 and 5p13.1 loci with Crohn’s disease in Japanese patients. J. Hum. Genet. 52, 575–583 (2007).
Davidson, S.I.*, Wu, X.*, Liu, Y., Wei, M., Danoy, P.A., Thomas, G. et al. Association of ERAP1, but not IL23R, with ankylosing spondylitis in a Han Chinese population. Arthritis Rheum. 60, 3263–3268 (2009).
Mori, M., Yamada, R., Kobayashi, K., Kawaida, R. & Yamamoto, K. Ethnic differences in allele frequency of autoimmune-diseaseassociated SNPs. J. Hum. Genet. 50, 264–266 (2005).
Brewerton, D. A., Hart, F. D., Nicholls, A., Caffrey, M., James, D. C. & Sturrock, R. D. Ankylosing spondylitis and HL-A 27. Lancet 1, 904–907 (1973).
Caffrey, M. F. & James, D. C. Human lymphocyte antigen association in ankylosing spondylitis. Nature 242, 121 (1973).
Schlosstein, L., Terasaki, P. I., Bluestone, R. & Pearson, C. M. High association of an HL-A antigen, W27, with ankylosing spondylitis. N. Engl. J. Med. 288, 704–706 (1973).
Karvonen, J. HL-A antigens in psoriasis with special reference to the clinical type, age of onset, exacerbations after respiratory infections and occurrence of arthritis. Ann. Clin. Res 7, 301–311 (1975).
Takano, M., Miyajima, T., Kiuchi, M., Ohmori, K. & Amemiya, H. Behçet disease and the HLA system. Tissue Antigens. 8, 95–99 (1976).
Hampe, J., Franke, A., Rosenstiel, P., Till, A., Teuber, M., Huse, K. et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 39, 207–211 (2007).
Ogura, Y., Bonen, D. K., Inohara, N., Nicolae, D. L., Chen, F. F., Ramos, R. et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature 411, 603–606 (2001).
UK IBD Genetics Consortium, Barrett, J. C., Lee, J. C., Lees, C. W., Prescott, N. J., Anderson, C. A. et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat. Genet. 41, 1330–1334 (2009).
Hugot, J. P., Chamaillard, M., Zouali, H., Lesage, S., Cézard, J. P., Belaiche, J. et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 411, 599–603 (2001).
Rioux, J. D., Xavier, R. J., Taylor, K. D., Silverberg, M. S., Goyette, P., Huett, A. et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 39, 596–604 (2007).
Parkes, M., Barrett, J. C., Prescott, N. J., Tremelling, M., Anderson, C. A., Fisher, S. A. et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat. Genet. 39, 830–832 (2007).
Dechairo, B. M., Zabaneh, D., Collins, J., Brand, O., Dawson, G. J., Green, A. P. et al. Association of the TSHR gene with Graves' disease: the first disease specific locus. Eur. J. Hum. Genet. 13, 1223–1230 (2005).
Lyons, P. A., Rayner, T. F., Trivedi, S., Holle, J. U., Watts, R. A., Jayne, D. R. et al. Genetically distinct subsets within ANCA-associated vasculitis. N. Engl. J. Med. 367, 214–223 (2012).
Suzuki, A., Yamada, R., Chang, X., Tokuhiro, S., Sawada, T., Suzuki, M. et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat. Genet. 34, 395–402 (2003).
Hreggvidsdottir, H. S., Noordenbos, T. & Baeten, D. L. Inflammatory pathways in spondyloarthritis. Mol. Immunol. 57, 28–37 (2014).
Diarra, D., Stolina, M., Polzer, K., Zwerina, J., Ominsky, M. S., Dwyer, D. et al. Dickkopf-1 is a master regulator of joint remodeling. Nat. Med. 13, 156–163 (2007).
Uderhardt, S., Diarra, D., Katzenbeisser, J., David, J. P., Zwerina, J., Richards, W. et al. Blockade of Dickkopf (DKK)-1 induces fusion of sacroiliac joints. Ann. Rheum. Dis. 69, 592–597 (2010).
Okada, Y., Wu, D., Trynka, G., Raj, T., Terao, C., Ikari, K. et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506, 376–381 (2014).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (30901321, 81373216, 81273283 and 81401355), and the National Basic Research Program (973 Program) of China (H. Xu, No.2014CB541802).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Wu, X., Chen, H. & Xu, H. The genomic landscape of human immune-mediated diseases. J Hum Genet 60, 675–681 (2015). https://doi.org/10.1038/jhg.2015.99
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2015.99
This article is cited by
-
Amphiphilic Triazine Polymer Derivatives as Antibacterial And Anti-atopic Agents in Mice Model
Scientific Reports (2019)
-
Human immune system diversity and its implications in diseases
Journal of Human Genetics (2015)