Abstract
Brown–Vialetto–Van Laere syndrome (BVVLS) is a very rare neurodegenerative disorder characterized by pontobulbar palsy and sensorineural hearing loss. Its mode of inheritance in affected families has usually been autosomal recessive, although autosomal dominant inheritance and incomplete penetrance have also been reported. Recently, C20orf54 was identified as a causative gene for BVVLS. Twelve different mutations have so far been identified in 10 patients affected with BVVLS or the related disorder Fazio Londe syndrome. Here, results of screening of C20orf54 in three unrelated BVVLS patients are reported. Four novel mutations that affect amino acid changes, p.Asn21Ser, p.Pro220His, p.Ala312Val and p.Gly375Asp, were identified in the patients. The causative nucleotide variations were not observed in 200 control individuals. One of the patients harbored compound heterozygous mutations, but only one mutated allele was observed in each of the two remaining patients.
Similar content being viewed by others
Introduction
Brown–Vialetto–Van Laere syndrome (BVVLS; MIM#211530) is a very rare neurodegenerative disorder characterized by pontobulbar palsy and sensorineural hearing loss.1, 2, 3, 4 Motor components of lower cranial nerves VII–XII are usually affected and lower motor neuron signs become evident. BVVLS is a progressive disorder that most often presents in the second decade of life, although reported age at onset of initial symptoms ranges from infancy to end of third decade.5, 6 Affected individuals are more likely to be female and this is thought to reflect a more severe and lethal presentation in males. Survival time is variable and ranges from a few years to several decades.4, 7 Respiratory compromise is the most common cause of demise. Sathasivam presented a comprehensive review of the disorder in 2008.4 Although first described in1894, only 58 cases had been described by the time of publication of that review.1, 4
In 2010, C20orf54 was identified as a BVVLS-causing gene after homozygosity mapping in a family with two affected individuals and the causative role of the gene was confirmed by identification of mutations therein in patients from six additional families.8 Genetic analysis of three additional unrelated patients, one of whom had been included in the earlier investigation, was later reported.9 In addition to confirming C20orf54 mutations in the patient previously studied, mutations in C20orf54 were found in one of the remaining patients. Mutations in C20orf54 were not observed in one of the three patients and the author ssuggested possible genetic heterogeneity for BVVLS.9 Most recently, mutations in the same gene were reported in two siblings affected with Fazio Londe syndrome (MIM#211500) and one unrelated individual affected with BVVLS.10 Fazio Londe syndrome has clinical features very similar to those of BVVLS, and they are considered by some to be a single disease entity.11, 12 The most distinguishable feature between the two disorders is absence of deafness in Fazio Londe syndrome-affected patients.4 C20orf54 is positioned on chromosome 20p13, and contains five exons. In all, mutations in C20orf54 have so far been reported in 10 unrelated individuals. All the patients harbored either homozygous or compound heterozygous mutations. This is consistent with the autosomal recessive mode of inheritance usually attributed to familial BVVLS prior to identification of a causative gene.4 Autosomal recessive inheritance was considered likely because affected pedigrees included multiple affected siblings born to unaffected parents.2, 3, 13, 14, 15 Here, we report the results of mutation screening of C20orf54 in three BVVLS-affected individuals. All three carried mutations in the gene. However, only one mutated allele was observed in two of the patients. The implications of this finding are discussed.
materials and Methods
This research was performed in accordance with the Declaration of Helsinki and with the approval of the ethics board of the University of Tehran. Participants and their guardians consented to participate after being informed of the nature of the research.
The three unrelated Iranian BVVLS patients were diagnosed based on clinical presentations, results of electrodiagnostic examinations and pure tone audiometry. Brain magnetic resonance imaging and systemic studies including chest X-ray imaging, abdominal ultrasonography and biochemical laboratory tests were also performed.
The exons and flanking intronic sequences of C20orf54 were amplified from DNA of leukocytes of three BVVLS-affected individuals. Amplicons were sequenced using the ABI Big Dye terminator chemistry and an ABI Prism 3700 instrument (Applied Biosystems, Foster City, CA, USA). Sequences were analyzed with the Sequencher software (Gene Codes Corporation, Ann Arbor, MI, USA). Reference sequences used were NC_000020.10,NM_033409.3 and NP_212134.3. Effects of variant sequences on splicing were predicted using NNsplice0.9 (http://www.fruitfly.org/seq_tools/splice.html) and GENSCAN (http://genes.mit.edu/GENSCAN.html). For determination of the extent of conservation of amino acids altered because of the nucleotide variations found, amino acid sequences from homologous proteins of other species were obtained from ENSEMBL (http://www.ensembl.org/index.html) and aligned using the Clustal W2.0 software (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Variations deemed to be possibly disease causing were screened in available family members of the patients and in 200 ethnically matched control individuals by an allele-specific PCR protocol. The structural consequences of these variations were assessed using the SIFT insilico tool (http://sift.jcvi.org/www/SIFT_seq_submit2.html).16 The sequences of all primers used in this study are available upon request.
Results
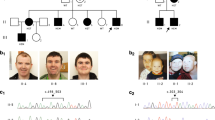
For all the patients, results of electromyography/nerve (EMG/NCV) conduction velocity were compatible with diagnosis of motor neuron disorder. There were evidences of active denervation and reinnervation in at least three segments of neuraxis, and normal nerve conductance was observed in the patients. Results of repeated nerve stimulation were negative. Pure tone audiometry revealed bilateral sensorineural hearing loss in the three individuals. Brain magnetic resonance imaging (MRI) and biochemical tests were unremarkable. Chest X-ray imaging and abdominal ultrasonography revealed no abnormalities. BVVLS was diagnosed in the patients. Their clinical features are summarized in Table 1. The age at onset of the earliest symptom in all three patients was in the second decade of life, and all were females born to non-consanguineous unaffected parents. Probands BVVL-100–3 and BVVL-101–3 did not have affected siblings. The parents of BVVL-102–4 reported that another daughter had developed symptoms like the proband and died at the age of 20 years, which was 18 months after presentation of earliest symptoms. In addition to the 10 previously reported relatively common sequence variations that are listed in the dbSNP database (http://www.ncbi.nlm.nih.gov/snp), four novel putative disease-causing mutations were observed (Figure 1). The four novel mutations affected amino acid changes. Case BVVL-100–3 carried compound heterozygous mutations, c.A62G and c.C935A, that, respectively, caused p.Asn21Ser and p.Ala312Val. Her parents and two unaffected siblings were heterozygous for one or the other mutation, suggesting segregation with disease status and autosomal recessive inheritance (Figure 1a). A c.C659A mutation causing p.Pro220His was observed in case BVVL-101–3, and a c.G1124A mutation causing p.Gly375Asp was found in BVVL-102–4 (Figures 1b and c). A second mutation was observed neither in BVVL-101–3 nor in BVVL-102–4. BVVL-101–3 inherited her mutated allele from her mother; her two unaffected brothers were not available for genetic screening (Figure 1b). BVVL-102–4 inherited her mutation from her father; her unaffected sister was homozygous for the wild-type allele (Figure 1c). Amino acids p.Asn21, p.Pro220 and p.Ala312 are well conserved in the homologous protein of distally related species (Table 2). Glycine at position 375 is substituted by alanine in some species; however, Gly>Ala is considered to be a conservative amino acid substitution. The observed mutation is non-conservative and causes substitution of an uncharged amino acid (Gly) by a negatively charged amino acid (Asp). All four amino-acid substitutions were predicted by the SIFT bioinformatics tool to be ‘not tolerated’. None of the putative disease-associated mutations was observed in the chromosomes of 200 control individuals.
BVVLS families and sequence chromatograms evidencing mutations. Genotypes of individuals genetically analyzed are indicated. Filled circles, BVVLS affected; open circles and squares, not affected. Chromatograms of family BVVL-100 (a) shows that the patient carries two mutations, each in the heterozygous state. Each of her parents carries only one mutated allele. Chromatogram of 101-3 (b) shows the patient's single mutation in the heterozygous state; her unaffected mother carries the same mutation in the heterozygous state. Chromatogram of 102-3 (c) shows the patient's single mutation in the heterozygous state; her unaffected father carries the same mutation in the heterozygous state. A full color version of this figure is available at the Journal of Human Genetics journal online.
Discussion
Four novel mutations in the known BVVLS-causing gene C20orf54 that affect amino acid changes p.Asn21Ser, p.Pro220His, p.Ala312Val and p.Gly375Asp are reported. Twelve different mutations in 10 BVVLS or Fazio Londe syndrome-affected individuals were previously reported (Figure 2). These 16 mutations are well spread out in the four amino acid coding exons of the gene; no mutation in exon 1 that codes 5'-noncoding sequences have been found. One mutation (causing p.Tyr213X) has been observed in two presumably unrelated patients.8, 10 C20orf54 codes a predicted transmembrane protein containing 469 amino acids and 11 transmembrane helices. The protein is the human ortholog of a rat riboflavin transporter that functions in the small intestine.17 The known missense mutations in C20orf54 are spread in the length of the coded protein; the affected amino acids are predicted to be positioned in transmembrane regions, intracellular loops or extracellular loops (Figure 2). Further mutation screening of C20orf54 and attention to genotype/phenotype correlations are expected to enhance understanding of the molecular basis and pathophysiology of BVVLS and related disorders.4
Schematic presentation of positions of known disease-causing mutations in C20orf54 and affected amino acids in the coded protein. Previously known mutations and novel mutations reported here, respectively, are shown with thin black arrows and thick unfilled arrows in gene (a) and coded protein (b). Protein model of the predicted transmembrane protein was obtained using TMHMM (http://www.cbs.dtu.dk/services/TMHMM-2.0/). Reference sequences NM_033409.3 and NP_212134.3 were used. In a, exons (E) are shown as boxes and introns (I) as thin lines between the boxes. The position of AUG initiation codon in exon 2 of the gene is shown.
The observation of only one mutated C20orf54 allele in two unrelated BVVLS patients is notable. All previously screened BVVLS patients with mutations in C20orf54 harbored two mutated alleles. Given the evolutionary conservation of the amino acids, it is also unlikely that the mutations causing p.Pro220His and p.Gly375Asp are very rare polymorphisms unrelated to disease status. P.Phe224Leu is a previously reported BVVLS-associated mutation that affects amino acids in close proximity to p.Pro220His, supporting the contention that a mutation affecting p.Pro220His can cause BVVLS. It is possible that a second mutation in patients BVVL-101–3 and BVVL-102–4 was missed. The second mutation may have been positioned deep within intronic sequences or in promoter or other non-coding regulatory regions that were not screened. Alternatively, the second mutation may have affected gene dosage changes. Nevertheless, it is striking that only one mutation in the coding region of C20orf54 was observed in two of three unrelated BVVLS patients screened in this study. The possibility that a single mutation in C20orf54 may with incomplete penetrance cause BVVLS should tentatively be considered. Careful and clinical neurologic examination of the parent (aged 58 years or older) of BVVL-101–3 and BVVL-102–4 who transmitted a mutation to an affected offspring confirmed the absence of disease signs. Variable expressivity and incomplete penetrance with respect to BVVLS have been discussed in the past because some relatives of BVVLS-affected individuals were observed who presented with only some of the clinical features of BVVLS.18, 19, 20, 21 In some families, mildly affected patients were diagnosed only after more severely affected sibs had been identified.20 Even possible autosomal dominant inheritance of BVVLS has been described in a few families.18, 19, 22, 23 The possibility that a single mutated allele in a gene can be cause of disease when usually two mutated alleles are causative has been considered for some neurologic disorders, such as for Parkinson’s disease with respect to PARKN and for some myopathies such as limb-girdle muscular dystrophy with respect to TTN.24, 25, 26, 27 The case of SOD1 mutations with respect to amyotrophic lateral sclerosis, which like BVVLS is a motor neuron disorder, is also interesting. SOD1 mutations are usually observed in amyotrophic lateral sclerosis families that exhibit autosomal dominant inheritance.28 However, a few mutations in this gene have been reported to be causative only in the homozygous state.29, 30 In any case, as association between C20orf54 and BBVLS has only recently been identified and the mode of inheritance of the disorder has not always been clear-cut, further screening of C20orf54 in BVVLS patients is warranted.
References
Brown, C. H. Infantile amyotrophic lateral sclerosis of the family type. J. Nerv. Ment. Dis. 19, 707–716 (1894).
Vialetto, E. Contributo alla forma ereditaria della paralisi bulbare progressive. Riv. Sper. Freniat. 40, 1–24 (1936).
Van Laere, J. [Familial progressive chronic bulbo-pontine paralysis with deafness. A case of Klippel-Trenaunay syndrome in siblings of the same family. Diagnostic and genetic problems]. Rev. Neurol. (Paris) 115, 289–295 (1966).
Sathasivam, S. Brown-Vialetto-Van Laere syndrome. Orphanet J. Rare Dis. 3, 9 (2008).
Sathasivam, S., O'Sullivan, S., Nicolson, A., Tilley, P. J., Shaw, P. J. Brown-Vialetto-Van Laere syndrome: case report and literature review. Amyotroph. Lateral Scler. Other Motor Neuron Disord. 1, 277–281 (2000).
Piccolo, G., Marchioni, E., Maurelli, M., Simonetti, F., Bizzetti, F., Savoldi, F. Recovery from respiratory muscle failure in a sporadic case of Brown-Vialetto-Van Laere syndrome with unusually late onset. J. Neurol. 239, 355–356 (1992).
Nemoto, H., Konno, S., Nomoto, N., Wakata, N., Kurihara, T. [A case of Brown-Vialetto-van Laere (BVVL) syndrome in Japan]. Rinsho. Shinkeigaku. 45, 357–361 (2005).
Green, P., Wiseman, M., Crow, Y. J., Houlden, H., Riphagen, S., Lin, J. P. et al. Brown-Vialetto-Van Laere syndrome, a ponto-bulbar palsy with deafness, is caused by mutations in c20orf54. Am. J. Hum. Genet. 86, 485–489 (2010).
Johnson, J. O., Gibbs, J. R., Van Maldergem, L., Houlden, H., Singleton, A. B. Exome sequencing in Brown-Vialetto-van Laere syndrome. Am. J. Hum. Genet. 87, 567–569 author reply 9-70 (2010).
Bosch, A. M., Abeling, N. G., Ijlst, L., Knoester, H., van der Pol, W. L., Stroomer, A. E. et al. and Fazio Londe syndrome is associated with a riboflavin transporter defect mimicking mild MADD: a new inborn error of metabolism with potential treatment. J. Inherit. Metab. Dis. 34, 159–164 (2011).
Dipti, S., Childs, A. M., Livingston, J. H., Aggarwal, A. K., Miller, M., Williams, C. et al. Brown-Vialetto-Van Laere syndrome; variability in age at onset and disease progression highlighting the phenotypic overlap with Fazio-Londe disease. Brain Dev. 27, 443–446 (2005).
McShane, M. A., Boyd, S., Harding, B., Brett, E. M., Wilson, J. Progressive bulbar paralysis of childhood. A reappraisal of Fazio-Londe disease. Brain 115 ((Part 6)), 1889–1900 (1992).
Boudin, G., Pepin, B., Vernant, J. C., Gautier, B., Gouerou, B. Cas familial de paralysie bulbo-pontine chronique progressive avec surdite. Rev. Neurol. (Paris) 124, 90–92 (1971).
Gallai, V., Hockaday, J. M., Hughes, J. T., Lane, D. J., Oppenheimer, D. R., Rushworth, G. Ponto-bulbar palsy with deafness (Brown-Vialetto-Van Laere syndrome). J. Neurol. Sci. 50, 259–275 (1981).
Malheiros, J. A., Camargos, S. T., Oliveira, J. T., Cardoso, F. E. A. Brazilian family with Brown-Vialetto-van Laere syndrome with autosomal recessive inheritance. Arq. Neuropsiquiatr. 65, 32–35 (2007).
Adzhubei, I. A., Schmidt, S., Peshkin, L., Ramensky, V. E., Gerasimova, A., Bork, P. et al. A method and server for predicting damaging missense mutations. Nat. Methods. 7, 248–249 (2010).
Yamamoto, S., Inoue, K., Ohta, K. Y., Fukatsu, R., Maeda, J. Y., Yoshida, Y. et al. Identification and functional characterization of rat riboflavin transporter 2. J. Biochem. 145, 437–443 (2009).
Summers, B. A., Swash, M., Schwartz, M. S., Ingram, D. A. Juvenile-onset bulbospinal muscular atrophy with deafness: Vialetta-van Laere syndrome or Madras-type motor neuron disease? J. Neurol. 234, 440–442 (1987).
Hawkins, S. A., Nevin, N. C., Harding, A. E. Pontobulbar palsy and neurosensory deafness (Brown-Vialetto-Van Laere syndrome) with possible autosomal dominant inheritance. J. Med. Genet. 27, 176–179 (1990).
Lombaert, A., Dom, R., Carton, H., Bruchler, J. M. Progressive ponto-bulbar palsy with deafness. A clinico-pathological study. Acta. Neurol. Belg. 76, 309–314 (1976).
Tavares, A. C. C., De Mattos, J. P., De Amarim, C. A. Donnier cochl6o-vestibulaires dans la scl6rose lat6rale amyotrophique (forme de Van Laere). Rev. Laryngol. Otol. Rhinol. 106, 375–378 (1985).
Van Laere, J. Over een newl geval van chosnische bulbopontiene paralysis met doofheid. Verh. Vlaan. Akad. Geneesk. Belg. 30, 288–308 (1967).
De Grandis, D., Passadore, P., Chinaglia, M., Brazzo, F., Ravenni, R., Cudia, P. Clinical features and neurophysiological follow-up in a case of Brown-Vialetto-Van Laere syndrome. Neuromuscul. Disord. 15, 565–568 (2005).
Bruggemann, N., Mitterer, M., Lanthaler, A. J., Djarmati, A., Hagenah, J., Wiegers, K. et al. Frequency of heterozygous Parkin mutations in healthy subjects: need for careful prospective follow-up examination of mutation carriers. Parkinsonism Relat. Disord. 15, 425–429 (2009).
Lesage, S., Lohmann, E., Tison, F., Durif, F., Durr, A., Brice, A. Rare heterozygous parkin variants in French early-onset Parkinson disease patients and controls. J. Med. Genet. 45, 43–46 (2008).
Udd, B., Vihola, A., Sarparanta, J., Richard, I., Hackman, P. Titinopathies and extension of the M-line mutation phenotype beyond distal myopathy and LGMD2J. Neurology 64, 636–642 (2005).
Penisson-Besnier, I., Hackman, P., Suominen, T., Sarparanta, J., Huovinen, S., Richard-Cremieux, I. et al. Myopathies caused by homozygous titin mutations: limb-girdle muscular dystrophy 2 J and variations of phenotype. J. Neurol. Neurosurg. Psychiatry 81, 1200–1202 (2010).
Ferrera, L., Caponnetto, C., Marini, V., Rizzi, D., Bordo, D., Penco, S. et al. An Italian dominant FALS Leu144Phe SOD1 mutation: genotype-phenotype correlation. Amyotroph. Lateral Scler. Other Motor Neuron Disord. 4, 167–170 (2003).
Al-Chalabi, A., Andersen, P. M., Chioza, B., Shaw, C., Sham, P. C., Robberecht, W. et al. Recessive amyotrophic lateral sclerosis families with the D90A SOD1 mutation share a common founder: evidence for a linked protective factor. Hum. Mol. Genet. 7, 2045–2050 (1998).
Jonsson, P. A., Backstrand, A., Andersen, P. M., Jacobsson, J., Parton, M., Shaw, C. et al. CuZn-superoxide dismutase in D90A heterozygotes from recessive and dominant ALS pedigrees. Neurobiol. Dis. 10, 327–333 (2002).
Acknowledgements
This work was supported by the Iran National Science Foundation. We thank the patients and their family members for consenting to participate in this study.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Dezfouli, M., Yadegari, S., Nafissi, S. et al. Four novel C20orf54 mutations identified in Brown–Vialetto–Van Laere syndrome patients. J Hum Genet 57, 613–617 (2012). https://doi.org/10.1038/jhg.2012.70
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2012.70
Keywords
This article is cited by
-
A Chinese pedigree with Brown-Vialetto-Van Laere syndrome due to two novel mutations of SLC52A2 gene: clinical course and response to riboflavin
BMC Medical Genetics (2019)
-
Disruption of Slc52a3 gene causes neonatal lethality with riboflavin deficiency in mice
Scientific Reports (2016)
-
Clinical presentation and outcome of riboflavin transporter deficiency: mini review after five years of experience
Journal of Inherited Metabolic Disease (2016)
-
The Brown-Vialetto-Van Laere and Fazio Londe syndrome revisited: natural history, genetics, treatment and future perspectives
Orphanet Journal of Rare Diseases (2012)
-
A Commentary on ‘Four novel C20ORF54 mutations identified in Brown–Vialetto–Van Laere syndrome patients.’
Journal of Human Genetics (2012)