Abstract
Fabry disease is a genetic disorder caused by deficient activity of lysosomal enzyme α-galactosidase A (GLA) and end-stage renal disease (ESRD) will be present after accumulation of glycosphingolipids within the kidney. Undiagnosed atypical variants of Fabry disease, which are limited to renal involvement, were found in several ESRD patient populations. On the other hand, unexpectedly high frequencies of male subjects having the c.196G>C nucleotide change (p.E66Q) showing low α-GLA activity have been reported on Japanese and Korean screening for Fabry disease. However, several evidences indicate the c.196G>C is not a pathogenic mutation but is a functional polymorphism. In the present study, high-throughput screening of serum GLA could successfully indentify two Fabry disease patients in a cohort consisted of 1080 male hemodialysis patients. Moreover, our serum assay was able to distinguish two patients with disease-causing genetic mutations (p.G195V and p.M296I) from eight functional variants that showed relatively decreased enzyme activity (p.E66Q). In conclusion, high-throughput serum enzyme assay distinctly identified disease-causing mutants and functional variants of GLA gene in Japanese male hemodialysis patients. In addition, our results underscore the high prevalence of not only undiagnosed Fabry patients but functional variants of p.E66Q among the ESRD population.
Similar content being viewed by others
Introduction
Fabry disease is an X-linked heritable disorder of glycosphingolipid metabolism caused by the deficient activity of the lysosomal enzyme α-galactosidase A (GLA) (Online Mendelian Inheritance in Man 301499).1 This disorder engenders progressive accumulation of glycosphingolipids such as globotriaosylceramide within tissues and organs, especially in the skin, nervous system, eye, heart and kidney. Typically in male hemizygotes, clinical manifestations caused by glycosphingolipid accumulation, such as acroparesthesias, hypohidrosis and corneal opacities, are observed during childhood. In the fourth or fifth decade of life, vascular involvement and subsequent cardiac hypertrophy and arrhythmia, proteinuria and renal dysfunctions, and brain stroke develop.2, 3 Recent reports have described atypical variants of Fabry disease that are limited to cardiac or renal involvement.4, 5, 6 As they show no classical manifestation during childhood, several variant Fabry disease patients are presumably undiagnosed. Although renal biopsy is the gold standard for the diagnosis of kidney diseases, not all end-stage renal disease (ESRD) patients receive renal biopsy before induction of dialysis. Instead, clinical diagnoses, such as chronic glomerulonephritis, diabetic nephropathy and hypertensive nephrosclerosis, would be assigned to ESRD patients by physicians. Therefore, it is possible that undiagnosed Fabry disease patients can be found in ESRD patients with other diagnosis of kidney disease.
The clinical introduction of enzyme replacement therapy (ERT) with recombinant GLA in 2001 enables us to treat patients with Fabry disease more specifically. ERT is effective for alleviating neuropathic pain, hypertrophic cardiomyopathy and sweat dysfunction, and for stabilizing kidney function.7, 8, 9, 10, 11, 12 This effectiveness encouraged us to proceed with screening studies of ESRD patients to identify Fabry patients who had never been suspected as having GLA deficiency. A systematic review by Linthorst et al.13 described the prevalence of Fabry disease in male dialysis patients in 10 studies as 0.33% (95% confidence interval 0.20–0.47%).13 To date, four screening studies for 1989 male Japanese dialysis patients have identified 12 Fabry patients (0.60% (95% confidence interval 0.26–0.94%)).14, 15, 16, 17 Three of them used plasma GLA assaying as the screening method;15, 16, 17 one employed a dried blood spot test.14 This study was designed to identify undiagnosed renal Fabry patients among ESRD patients using a high-throughput screening system, which allowed us to measure GLA activity of serum samples distributed in 96-well plates within 4 h. We also measured GLA activity of peripheral leukocytes to confirm the diagnosis and to examine genetic mutations in the GLA gene (GLA) further.
Materials and methods
Study population
This study examined 1080 male ESRD patients who were treated by maintenance hemodialysis at 14 dialysis centers in Tokyo area, none of whom had been diagnosed previously as having Fabry disease. This study was approved by the Institutional Review Board (IRB) and the Ethics Committee for Human Genome Study in The University of Tokyo and Meiji Pharmaceutical University. As no dialysis center has its own IRB, IRB in The University of Tokyo reviewed the study protocol upon the request from each dialysis center. Written informed consent was obtained from each participant at the time of recruitment.
Measurement of serum and leukocyte GLA activity
Serum samples were obtained at the start of dialysis by immediate centrifuge (3000 r.p.m. × 10 min) and kept at 4 °C for up to 24 h. Then serum samples were further kept at −30 °C until GLA activity measurement. GLA activity was measured using a substrate solution containing 5 mmol l−1 4-methylumbelliferyl α-D-galactopyranoside (Calbiochem, La Jolla, CA, USA) as a substrate and 117 mmol l−1 N-acetyl-D-galactosamine (Sigma-Aldrich Corp, St Louis, MO, USA) as a specific inhibitor of N-acetylgalactosaminidase in 0.1 mol l−1 citrate–phosphate butter, pH 4.6.18 In the first screening, 20 μl of serum was mixed with 40 μl of the substrate solution in a 96-well plate. The mixture was incubated at 37 °C for 4 h. Then the reaction was stopped by adding 200 μl of 0.2 mol l−1 glycine buffer, pH 10.7, and the released 4-methylumbelliferone was measured using a Wallac 1420 ARVO MX Multilabel Counter (PerkinElmer Inc., Waltham, MA, USA) at excitation and emission wavelengths of 355 and 460 nm, respectively. When the serum GLA activity was lower than 1.5 nmol h−1 ml−1, the leukocyte GLA activity was measured. In the second examination, 10 μl of leukocyte homogenate (10 μg protein) was mixed with 40 μl of the substrate solution in a 1.5-ml microtube. The mixture was incubated at 37 °C for 30 min. Then, the reaction was stopped by adding 950 μl of 0.2 mol l−1 glycine buffer, pH 10.7, and the released 4-methylumbelliferone was measured using a spectrofluorometer (F2700; Hitachi Ltd, Tokyo, Japan) at excitation and emission wavelengths of 365 and 450 nm, respectively.
Genome DNA analysis of the GLA gene
Genomic DNA was purified from leukocytes with a Wizard Genomic DNA Purification Kit (Promega Corp., Madison, WI, USA) according to the manufacturer’s instructions. Seven exons of the GLA gene were amplified by PCR using appropriate primers.19 The PCR protocol consists of 2 min at 94 °C, then 35 cycles of 15 s at 94 °C, 30 s at 55 °C, 40 s at 68 °C, and 1 min at 68 °C with KOD-plus DNA polymerase (Toyobo Co. Ltd, Osaka, Japan). The PCR fragment was directly sequenced.
Structural modeling of mutant GLA proteins
Structural modeling of mutant GLA proteins was performed using molecular modeling software TINKER, as described previously.20 The crystal structure of human GLA (PDB: 1R46) was used as a template, and energy minimization was performed. The root-mean-square gradient value was set at 0.05 kcal mol−1 per Å. Coloring of the affected atoms in the three-dimensional structure of GLA was performed based on the distances between the wild-type and mutant ones.
Results
Patients
This study examined 1080 male ESRD patients treated by maintenance hemodialysis at 14 dialysis centers in Tokyo area. The causes of ESRD described in the medical records were chronic glomerulonephritis (n=376, 34.8%), diabetic nephropathy (n=372, 34.4%), nephrosclerosis (n=140, 13.0%), polycystic kidney disease (n=47, 4.4%) and miscellaneous other conditions (n=54, 5.0%). Also, 91 patients (8.4%) were described as ESRD with unknown etiology. Their age was 63.4±13.0 (mean±s.d.) years at enrollment. They had started to receive dialysis at 55.1±15.7 years.
Measurement of GLA activity
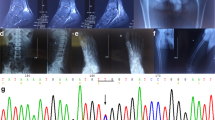
A preliminary study was performed to determine the cutoff point of serum GLA activity in this study. The serum GLA activities of 32 apparently normal subjects and 54 Fabry patients who had already been diagnosed were, respectively, 4.6±1.6 (mean±s.d.) and 0.1±0.1 nmol h−1 ml−1. Considering the result (mean−2 s.d.=1.4 nmol h−1 ml−1), we determined the cutoff point as 1.5 nmol h−1 ml−1 to avoid a false-negative case. Analysis with these 86 individuals revealed that this cutoff point provided highly accurate diagnosis of Fabry disease (sensitivity 100%, specificity 100%) (Figure 1). Figure 2 shows the distribution of serum GLA activity in the enrolled ESRD patients. The first screening revealed that the average of GLA activity of all the subjects was 4.6±1.3 nmol h−1 ml−1; 10 of the subjects showed enzyme activity lower than 1.5 nmol h−1 ml−1. Leukocyte GLA activity in these 10 patients was subsequently measured to confirm the Fabry disease diagnosis (Table 1). The average of leukocyte GLA activity of 33 apparently normal subjects was 49±20 nmol h−1 per mg protein. Two patients exhibited remarkably low GLA activity and were diagnosed as having Fabry disease (Cases 1 and 2). Notably, they were diagnosed as ESRD because of chronic glomerulonephritis when they started dialysis therapy. Case 1 was complicated with severe hypertrophic cardiomyopathy and arrhythmia (frequent premature ventricular contraction). Magnetic resonance imaging of the brain revealed multiple infarction. One brother died suddenly at the age of 37 years; another brother suffered from sick sinus syndrome. Case 2 was complicated with moderate left ventricular hypertrophy, but had no remarkable family history (Figure 3). None of these two patients showed any typical symptoms of classical Fabry disease, such as abnormal sweating, angiokeratoma and cornea verticillata. Eight other patients had residual enzyme activity of about 24–65% of the normal control mean (Cases 3–10).
Identification of gene mutation in GLA gene
Direct sequencing was conducted on all the exons of the GLA gene with the genomic DNA of the 10 patients who were selected by the first screening (Table 1 and Figure 4). A missense mutation of p.G195V was found in Case 1. Case 2 had another missence mutation of p.M296I. These had been identified in several patients with variant form of Fabry disease.4, 17, 21 The others (Cases 3–10) had a p.E66Q allele that had been reported previously as a functional polymorphism (rs28935191).22 Notably, serum GLA activity of these p.E66Q patients were higher than the p.G195V and p.M296I patients (Figure 5).
Prediction of structural changes caused by amino-acid substitutions
We constructed three-dimensional models of the GLA proteins with amino-acid substitutions attributable to p.G195V, p.M296I and p.E66Q. Coloring of the atoms affected by the amino-acid substitutions is shown in Supplementary Figure 6 (Supplementary information is available at Journal of Human Genetics’ website at the end of the article and before the references). In these cases, the predicted structural changes caused by the amino-acid substitutions are small and are localized on the molecular surface far from the active site.
Discussion
We conducted large-scale screening of Fabry disease with 1080 Japanese male ESRD patients using a high-throughput measurement of serum GLA activity followed by leukocyte GLA assaying, and successfully identified two disease-causing mutants and eight functional variants of the GLA gene. In the first screening of serum GLA measurement, 10 patients showed low GLA activity. The second test of leukocyte enzyme assay confirmed the diagnosis of Fabry disease with two patients whose GLA activity was almost deficient. They had genetic mutations of p.G195V in exon 4 and p.M296I in exon 6, which were identified in some patients with variant form of Fabry disease exhibiting renal or cardiac involvement.4, 17, 21
The incidence of classic Fabry disease has been estimated as 1 in 50 000 males.23 Dutch metabolic diagnostic centers reported birth prevalence of 1 in 238 000 in male newborns.24 However, Spada et al.25 found high incidence of later-onset Fabry disease (that is, cardiac and renal variants) by newborn screening. They reported that the incidence of GLA deficiency was about 1 in 3100, with an 11:1 ratio of patients with the variant: classic phenotypes. However, the results of screening studies of ‘high risk’ populations such as ESRD and hypertrophic cardiomyopathy showed higher incidence.13 The calculated prevalence of Fabry disease in this study was lower than that reported in previous studies that examined Japanese dialysis patients (0.19 vs 0.60%).14, 15, 16, 17
The difference is probably derived from the diagnostic criteria of Fabry disease. It should be addressed that previous studies included subjects with the p.E66Q allele as a ‘renal variant’ of Fabry disease.14, 15, 17, 26 However, Lee et al.22 reported that the allele frequency of p.E66Q in Korean individuals was remarkably higher (1.046% (95% confidence interval, 0.458–1.634%)) than the prevalence of Fabry disease. They also demonstrated that p.E66Q was a functional polymorphism rather than a pathogenic mutation using COS-7 cells overexpressing α-GLA harboring p.E66Q. More recently, we clearly demonstrated that p.E66Q was a functional variant and could be false negatively identified as Fabry disease because of its instability.27 Biochemical, pathological and structural studies revealed subjects harboring the p.E66Q exhibited relatively high residual enzyme activity in white blood cells as also shown in this study, and that there was no accumulation of globotriaosylceramide in cultured fibroblasts or an increased level of plasma globotriaosylsphingosine in these subjects. Of note, we also demonstrated the GLA enzyme in plasma from subjects harboring the p.E66Q enzyme decreased after incubation for 4 h at 37 °C, indicating the GLA with the E66Q amino-acid-substitution is more unstable and therefore the p.E66Q subject can be false positively detected as Fabry disease.27 On the other hand, the first screening using serum samples in the present study detected relatively but significantly higher GLA activity in eight p.E66Q patients than two disease-causing mutants (Table 1, Figure 5). Although the values were lower than the cutoff (1.5 nmol h−1 ml−1), this result indicates that serum assay of GLA activity can distinguish functional polymorphisms including p.E66Q from pathogenic mutations when a proper cutoff value is determined. It must be stated that not all the p.E66Q patients were identified in this study, because genetic analysis was conducted in only 10 patients who showed low-serum GLA activity. In addition, only two Fabry disease patients (positive control) were identified in this study. Further investigation with larger sample size is necessary to confirm the performance of serum GLA activity measurement as a screening method for Fabry disease.
To date, more than 500 mutations have been identified in the GLA gene.28 In this study, two disease-causing mutations of p.G195V and p.M296I were identified. The predicted structural change in this mutant protein is small and is located on the molecular surface far from the active site. The degree of structural change in these mutants is not large compared with that in the case of p.E66Q. However, even such minor conformational change is expected to be detected by the endoplasmic reticulum’s quality control system and the mutant GLA proteins might be degraded before transportation to lysosomes in these mutants, although subjects having these mutant alleles exhibit a mild phenotype of Fabry disease.20, 29
As described above, Fabry patients of variant type lack specific symptoms in childhood and show variable clinical expression at later time points. Therefore, such patients would be misdiagnosed and would lose the opportunity of receiving ERT, which is expected to be especially effective in less severely affected Fabry patients. ERT was not able to stabilize renal function in chronic kidney disease patients with severe proteinuria or a glomerular filtration rate of lower than 60 ml min−1.30 This study aimed at finding undiagnosed Fabry patients who were suffering from ESRD. Although ERT might not reverse far advanced renal failure, dialysis patients will have some benefits. ESRD patients are very frequently complicated with severe cardiovascular diseases.31 Fabry patients on dialysis would suffer from both primary Fabry-related cardiac disease and cardiovascular disease derived from ESRD. Another more important advantage is identification of undiagnosed patients in related family members. Neonatal mass screening of Fabry disease will be difficult in terms of ethical and cost-effective issues.25 Therefore, expanding screening from the ESRD population might be a reasonable strategy to identify Fabry patients' ancestry.
In conclusion, two undiagnosed variant type of Fabry patients and eight subjects with a functional polymorphism were identified in male Japanese hemodialysis patients using our high-throughput serum screening. Serum GLA activity was able to distinguish disease-causing mutants from a functional polymorphism (p.E66Q). Our result addressed the high prevalence of undiagnosed Fabry patients in ESRD population and the necessity of better strategy for finding variant Fabry patients in the stage, when they would have a benefit by ERT.
References
Brady, R.O., Gal, A.E., Bradley, R.M., Martensson, E., Warshaw, A.L., Laster, L. Enzymatic defect in Fabry's disease. Ceramidetrihexosidase deficiency. N. Engl. J. Med. 276, 1163–1167 (1967).
Desnick, R.J., Ioannou, Y., Eng, C.M. in The Metabolic and Molecular Bases of Inherited Disease. Scriver C.R., Beaudet A.L., Sly W.S. & Valle D. (eds). 3373–3374 (McGraw-Hill: New York, 2001).
Branton, M.H., Schiffmann, R., Sabnis, S.G., Murray, G.J., Quirk, J.M., Altarescu, G. et al. Natural history of Fabry renal disease: influence of alpha-galactosidase A activity and genetic mutations on clinical course. Medicine 81, 122–138 (2002).
Nakao, S., Takenaka, T., Maeda, M., Kodama, C., Tanaka, A., Tahara, M. et al. An atypical variant of Fabry's disease in men with left ventricular hypertrophy. N. Engl. J. Med. 333, 288–293 (1995).
Rosenthal, D., Lien, Y.H., Lager, D., Lai, L.W., Shang, S., Leung, N. et al. A novel alpha-galactosidase a mutant (M42L) identified in a renal variant of Fabry disease. Am. J. Kidney Dis. 44, e85–e89 (2004).
von Scheidt, W., Eng, C.M., Fitzmaurice, T.F., Erdmann, E., Hübner, G., Olsen, E.G. et al. An atypical variant of Fabry's disease with manifestations confined to the myocardium. N. Engl. J. Med. 324, 395–399 (1991).
Hughes, D.A., Elliott, P.M., Shah, J., Zuckerman, J., Coghlan, G., Brookes, J. et al. Effects of enzyme replacement therapy on the cardiomyopathy of Anderson-Fabry disease: a randomised, double-blind, placebo-controlled clinical trial of agalsidase alfa. Heart 94, 153–158 (2008).
Schiffmann, R., Floeter, M.K., Dambrosia, J.M., Gupta, S., Moore, D.F., Sharabi, Y. et al. Enzyme replacement therapy improves peripheral nerve and sweat function in Fabry disease. Muscle Nerve. 28, 703–710 (2003).
Schiffmann, R., Ries, M., Timmons, M., Flaherty, J.T., Brady, R.O. Long-term therapy with agalsidase alfa for Fabry disease: safety and effects on renal function in a home infusion setting. Nephrol. Dial. Transplant. 21, 345–354 (2006).
Banikazemi, M., Bultas, J., Waldek, S., Wilcox, W.R., Whitley, C.B., McDonald, M. et al. Agalsidase-beta therapy for advanced Fabry disease: a randomized trial. Ann. Intern. Med. 146, 77–86 (2007).
Schiffmann, R., Kopp, J.B., Austin, H.A., Sabnis, S., Moore, D.F., Weibel, T. et al. Enzyme Replacement Therapy in Fabry Disease. JAMA 285, 2743–2749 (2001).
West, M., Nicholls, K., Mehta, A., Clarke, J.T., Steiner, R., Beck, M. et al. Agalsidase alfa and kidney dysfunction in Fabry disease. J. Am. Soc. Nephrol. 20, 1132–1139 (2009).
Linthorst, G.E., Bouwman, M.G., Wijburg, F.A., Aerts, J.M., Poorthuis, B.J., Hollak, C.E. Screening for Fabry disease in high-risk populations: a systematic review. J. Med. Genet. 47, 217–222 (2010).
Fujii, H., Kono, K., Goto, S., Onishi, T., Kawai, H., Hirata, K. et al. Prevalence and cardiovascular features of Japanese hemodialysis patients with Fabry disease. Am. J. Nephrol. 30, 527–535 (2009).
Tanaka, M., Ohashi, T., Kobayashi, M., Eto, Y., Miyamura, N., Nishida, K. et al. Identification of Fabry's disease by the screening of alpha-galactosidase A activity in male and female hemodialysis patients. Clin. Nephrol. 64, 281–287 (2005).
Ichinose, M., Nakayama, M., Ohashi, T., Utsunomiya, Y., Kobayashi, M., Eto, Y. Significance of screening for Fabry disease among male dialysis patients. Clin. Exp. Nephrol. 9, 228–232 (2005).
Nakao, S., Kodama, C., Takenaka, T., Tanaka, A., Yasumoto, Y., Yoshida, A. et al. Fabry disease: detection of undiagnosed hemodialysis patients and identification of a ‘renal variant’ phenotype. Kidney Int. 64, 801–807 (2003).
Mayes, J.S., Scheerer, J.B., Sifers, R.N., Donaldson, M.L. Differential assay for lysosomal alpha-galactosidases in human tissues and its application to Fabry's disease. Clin. Chem. Acta. 112, 247–251 (1981).
Takata, T., Okumiya, T., Hayashibe, H., Shimmoto, M., Kase, R., Itoh, K. et al. Screening and detection of gene mutations in Japanese patients with Fabry disease by non-radioactive single-stranded conformation polymorphism analysis. Brain Dev. 19, 111–116 (1997).
Saito, S., Ohno, K., Sese, J., Sugawara, K., Sakuraba, H. Prediction of the clinical phenotype of Fabry disease based on protein sequential and structural information. J. Hum. Genet. 55, 175–178 (2010).
Nakagawa, N., Maruyama, H., Ishihara, T., Seino, U., Kawabe, J., Takahashi, F. et al. Clinical and genetic investigation of a Japanese family with cardiac fabry disease. Identification of a novel alpha-galactosidase A missense mutation (G195V). Int. Heart J. 52, 308–311 (2011).
Lee, B.H., Heo, S.H., Kim, G.H., Park, JY., Kim, W.S., Kang, D.H. et al. Mutations of the GLA gene in Korean patients with Fabry disease and frequency of the E66Q allele as a functional variant in Korean newborns. J. Hum. Genet. 55, 512–517 (2010).
Meikle, P.J., Hopwood, J.J., Clague, A.E., Carey, W.F. Prevalence of Lysosomal Storage Disorders. JAMA 281, 249–254 (1999).
Poorthuis, B.J., Wevers, R.A., Kleijer, W.J., Groener, J.E., de Jong, J.G., van Weely, S. et al. The frequency of lysosomal storage diseases in The Netherlands. Hum. Genet. 105, 151–156 (1999).
Spada, M., Pagliardini, S., Yasuda, M., Tukel, T., Thiagarajan, G., Sakuraba, H. et al. High incidence of later-onset fabry disease revealed by newborn screening. Am. J. Hum. Genet. 79, 31–40 (2006).
Shimotori, M., Maruyama, H., Nakamura, G., Suyama, T., Sakamoto, F., Itoh, M. et al. Novel mutations of the GLA gene in Japanese patients with Fabry disease and their functional characterization by active site specific chaperone. Hum. Mutat. 29, 331 (2008).
Togawa, T., Tsukimura, T., Kodama, T., Tanaka, T., Kawashima, I., Saito, S. et al. Fabry disease: biochemical, pathological and structural studies of the alpha-galactosidase A with E66Q amino acid substitution. Mol. Genet. Metab. (2012).
Saito, S., Ohno, K., Sakuraba, H. Fabry-database.org: database of the clinical phenotypes, genotypes and mutant alpha-galactosidase A structures in Fabry disease. J. Hum. Genet. 56, 467–468 (2011).
Sugawara, K., Ohno, K., Saito, S., Sakuraba, H. Structural characterization of mutant alpha-galactosidases causing Fabry disease. J. Hum. Genet. 53, 812–824 (2008).
Germain, D.P., Waldek, S., Banikazemi, M., Bushinsky, D.A., Charrow, J., Desnick, R.J. et al. Sustained, long-term renal stabilization after 54 months of agalsidase beta therapy in patients with Fabry disease. J. Am. Soc. Nephrol. 18, 1547–1557 (2007).
Foley, R.N., Parfrey, P.S., Sarnak, M.J. Clinical epidemiology of cardiovascular disease in chronic renal disease. Am. J. Kidney Dis. 32, S112–S119 (1998).
Acknowledgements
The authors thank Drs Hiromi Shimoyama, Toshimasa Takahashi, Mitsumine Fukui, Hyoe Tomita, Akiko Ogura, Takashi Ozawa, Katsufumi Sakata, Miho Enomoto, Takako Suzuki, Junichiro Mera and Naoto Fujizuka for providing samples. This work was partly supported by Grants from the Japan Society for the Promotion of Science (ID: 23659527 and 21390314, HS and ID: 24390212, EN); the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (ID: 09-15, HS); the Program for Research on Intractable Diseases of Health and Labor Science Research Grants (HS); the JAPS Asia/Africa Scientific Platform Program (HS); and the High-Tech Research Center Project of the Ministry of Education, Culture, Sports, Science and Technology of Japan (ID: S0801043, HS).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Doi, K., Noiri, E., Ishizu, T. et al. High-throughput screening identified disease-causing mutants and functional variants of α-galactosidase A gene in Japanese male hemodialysis patients. J Hum Genet 57, 575–579 (2012). https://doi.org/10.1038/jhg.2012.68
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2012.68
Keywords
This article is cited by
-
The Ckd. Qld fabRy Epidemiology (aCQuiRE) study protocol: identifying the prevalence of Fabry disease amongst patients with kidney disease in Queensland, Australia
BMC Nephrology (2020)
-
Identifying Fabry patients in dialysis population: prevalence of GLA mutations by renal clinic screening, 1995–2019
Journal of Nephrology (2020)
-
Fabry disease in children: a federal screening programme in Russia
European Journal of Pediatrics (2017)
-
Prevalence of Fabry disease in dialysis patients: Japan Fabry disease screening study (J-FAST)
Clinical and Experimental Nephrology (2016)
-
Fabry disease: experience of screening dialysis patients for Fabry disease
Clinical and Experimental Nephrology (2014)