Abstract
Dravet syndrome (DS) or severe myoclonic epilepsy of infancy is an intractable epileptic syndrome that is caused by mutations in the neuronal voltage-gated sodium channel α1 subunit gene SCN1A. We investigated SCN1A mutations in 63 Chinese patients with DS and analyzed its inheritance. Genomic DNA was extracted from peripheral blood lymphocytes of DS patients and their available parents. The SCN1A open reading frame sequence was analyzed by PCR–DNA sequencing and multiple ligation-dependent probe amplication (MLPA). If the mutation was de novo, we used allele-specific PCR (AS-PCR) to determine the parental origin. Of the 63 patients examined, 49 unrelated patients had SCN1A mutations. The mutation rate was 77.8% (49 of 63), in which 61.2% (30 of 49) were truncation mutations. The mutations included 19 missense mutations, 14 frame-shift mutations, 6 nonsense mutations, 8 splice-site mutations. Through MLPA analysis, deletions or duplications of large fragments accounted for 12.5% (2 of 16) in PCR-sequencing-negative patients. By testing parents for the mutation, 40 mutations were found to be de novo and one mutation was inherited from a mother who was mosaic for a mutation. By AS-PCR analysis in 12 patients with de novo mutations, 10 were confirmed paternal in origin and 2 were maternal in origin. Thirty of the SCN1A mutations reported here have not been previously reported. Approximately 80% of Chinese DS patients have SCN1A mutations. MLPA analysis was essential for PCR-sequencing-negative patients. The majority of SCN1A mutations were de novo, most of which were paternal origin.
Similar content being viewed by others
Introduction
Dravet syndrome (DS) or severe myoclonic epilepsy of infancy is an intractable epileptic syndrome beginning with febrile hemiclonic or generalized tonic–clonic seizures in the first year of life. Subsequently, multiple seizure types evolve, such as myoclonic, focal or absence seizures. Psychomotor developmental is typically delayed after 1 year. Ataxia and pyramidal signs were observed in some patients. The seizures are refractory to antiepileptic drugs.1, 2 DS had a poor long-term prognosis. The majority of DS patients still had attacks in their adulthood, with moderate or severe mental retardation.3, 4 Mutations in the voltage-gated sodium channel α1 subunit gene SCN1A is the major pathogenic cause of DS. Analysis of SCN1A mutations has become a diagnostic test for DS. To date, more than 400 SCN1A mutations have been identified in DS.5, 6, 7, 8
We have carried out SCN1A mutation analysis in 63 Chinese patients with DS using PCR-sequencing and multiple ligation-dependent probe amplification (MLPA). For patients found to be positive for an SCN1A mutation, we then carried out mutation analysis in their parents, when available, to investigate the inheritance of the mutation. In the case of de novo mutations, allele-specific PCR (AS-PCR) was used to determine the chromosome of origin of the de novo mutation.
Patients And Methods
Patients
Sixty-three patients were consistent with the clinical features of DS. They were examined at the Peking University First Hospital from February 2005 to April 2009. Written informed consent was obtained from all participants. The clinical diagnosis of DS were as follows:1, 9 (1) seizure onset within 1 year of age (around 6 months), first event is often febrile seizures (FS); (2) normal early development; (3) prolonged generalized or hemiclonic seizures, often triggered by fever; (4) multiple seizure types (myoclonic, focal, atypical absences) evolve after the second year of life; (5) persistence of seizure sensitivity to fever; (6) psychomotor slowing after the second year of life, ataxia and pyramidal signs may evolve; (7) interictal electroencephalography were normal in the first year of life, followed by generalized, focal or multifocal discharges; and (8) pharmacoresistance. The fifty normal controls had no history of FS and epilepsy.
DNA isolation and bioinformatics analysis
Genomic DNA was extracted from peripheral blood lymphocytes of the patients and their available parents by a simple salting-out procedure.10 The 26 exons of SCN1A were amplified by PCR using primers as previously described.11 Samples with mutations were sequenced in both directions from two independent PCR products. The corresponding segments of the mutation-positive patients’ parents were examined to determine from which parent the mutation was inherited. The protein domains were analyzed using ScanProsite (http://www.expasy.org/, PRO_0000048489). Sequence alignments of the sodium channel α subunits in vertebrate and invertebrate species were performed using ClustalW (http://www.ebi.ac.uk/clustalw/). We used the SCN1A mutation database (HGMD, http://www.hgmd.cf.ac.uk/ac/all.php) and related data to examine whether these mutations are novel.
MLPA
If no mutation was identified in the DS patient by PCR-sequencing, we used MLPA kit P137 (MRC-Holland, Amsterdam, the Netherlands) to screen for large deletions or duplications. The MLPA reactions were carried out following the instructions provided by the manufacturer. Finally, the PCR products were separated by capillary electrophoresis using an ABI 3100 automated sequencer and size standards (Perkin-Elmer Applied Biosystems, Foster City, CA, USA). We used Genescan analysis software (version 3.7) and Genotype software (version 3.6) to analyze these data, which we then exported to a Microsoft Excel spreadsheet. The resulting values were approximately 1 for every wild-type peak, <0.7 for heterozygous deletions and >1.3 for heterozygous duplications.
AS-PCR
Referring to the NCBI single-nucleotide polymorphism (SNP) database and SNPs identified in this study, we selected SNPs in SCN1A, which were close to our identified mutations. We then designed primers to amplify the corresponding SNP-containing fragments in the probands and their parents. When the proband’s SNP was heterozygous and one of his or her parent’s SNP was homozygous or both were different homozygous, we were able to determine the SNP origin of the proband. The primers are shown in Supplementary Table 1.
For AS-PCR, one or two pairs of allele-specific primers were designed based on the informative SNPs, which were located at the 3′-terminus of primers. The amplified fragments contained the mutation locus. The nucleotides at the 3′-terminus of the primer were complementary to one allele, but were mismatched to another allele. Under carefully controlled conditions, a primer with its terminal 3′-nucleotide mismatched will not function properly, and no amplification will occur from another allele. Subsequently, we could detect whether the alleles we amplified were wild type or mutant by sequencing. AS-PCR primers are listed in Supplementary Table 2.
Microsatellite analysis of informative families
The 12 informative families were tested to confirm family relationships using five highly polymorphic microsatellite markers. These were D5S424, D6S289, D13S175, D15S117 and D16S3136. PCRs were carried out according to the manufacturer's instructions (ABI-Perkin-Elmer). The reverse primer of each pair was labeled with HEX, NED or FAM. PCR products were analyzed on ABI 3730 automated sequencer and Genotype software (GeneMapper 3.0).
Results
SCN1A mutation
Patients consistent with the clinical diagnosis of DS were selected for SCN1A mutation analysis. In 63 patients, 49 had SCN1A mutations. The mutation rate was 77.8% (49 of 63), of which more than half (30 of 49, 61.2%) were truncation mutations.
PCR-sequencing analysis. Most mutations (47 of 49, 95.9%) were identified by PCR-sequencing (see Table 1 and Figure 1). In all, 47 patients had 45 different mutations, of which R101W and A1783T were recurrent in two unrelated cases, respectively. The recurrent mutations accounted for 4.1% (2 of 49) in the total mutations. All the mutations were heterozygous, with missense mutations being the most prevalent (19 of 49, 38.8%), followed by frame-shift mutations (14 of 49, 28.6%), splice-site mutations (8 of 49, 16.3%) and nonsense mutations (6 of 49, 12.2%). The mutations were scattered in different exons. Mutations in the exon 26 were the most prevalent (7 of 49, 14.3%), followed by exons 2 and 19 (each 10.2%, 5 of 49), and then exons 10, 15 and 21 (each 8.2%, 4 of 49). The remaining mutations were scattered in the other 20 exons.
Altogether, 33 mutations were caused by base substitutions accounting for 67.3%, of which 24 mutations (G>A, 13; C>T, 6; T>C, 4; A>G, 1) may be due to nucleotide deamination and 3 mutations due to oxidatively damaged bases (G>T, 2; C>A, 1). The other 14 mutations were frame shift accounting for 28.6%, of which 8 were caused by deletions, 4 by insertions, and 2 mutations were of both insertions and deletions in the same locus. In the eight deletion mutations, five were caused by depurination (5 of 8, 62.5%), and the other three in the following ways: one case deleting ‘TT’; one deletion from two repeat ‘AAACAAAC’ to one copy of ‘AAAC’; and one case containing ‘ATTCAGAGA’ deleted to ‘ATTGAGA’. In four insertion mutations, three were specific sequence repeats: one case was from ‘GACC’ to ‘GACCGACC’; one case from five repeats ‘ATATATATAT’ to six repeats ‘ATATATATATAT’; one case from ‘AAGTA’ to ‘AAG TAAAGTA’; and one case from ‘T’ to ‘TT’. The frequency of mutations caused by deamination (49%, 24 of 49) was significantly higher than that caused by other factors such as oxidation (6.12%, 3 of 49) and depurination (10.2%, 5 of 49).
Missense mutations located in the important functional domains accounted for nearly half (8 of 19, 42.1%) of the total missense mutations. Conservative analysis showed that I91T, V1390M, T1210K and G1586E were located in the moderate conservative region. The remaining missense mutations were highly conservative. These four mutations were all involved with charge property changes of amino acids. Except R393H and L1287P, the other 17 missense mutations referred to the changes of the charge state of the amino acid. However, R393H was located in the pore region, and L1287P in the highly conserved transmembrane domain.
MLPA analysis. Direct sequencing of PCR-DNA screening of SCN1A mutations was negative in 16 DS patients and these patients were further studied by MLPA analysis (see Table 1 and Figure 2). In the case number 48, there were eight-exon duplications, including exons 3, 4, 5, 6, 7, 8, 9 and 10 extending about 12 Kb. In the case number 49, the patient had two-exon deletions, which included exons 22 and 23 spanning ∼3 Kb. SCN1A deletion and duplication of large fragments accounted for 4.1% (2 of 49) of the total patients with mutations, and accounting for 12.5% (2 of 16) of PCR-DNA sequencing-negative cases.
Origin of mutation analysis
Source of mutations. For the 49 children with SCN1A mutations, we also carried out mutation analysis in their parents with PCR-DNA sequencing and MLPA to detect the mutation origin. It was revealed that 40 mutations were de novo and only one mutation (case number 5, R101Q) was inherited from her mosaic mother. As for the case number 47, DNA of both his parents was not available, so we could not further analyze the source of the mutation. In the remaining seven patients, none of their available one parent carried these mutations (Table 1).
In case number 5 with the mutation p.R101Q (c.302G>A), her sister also carried this mutation. PCR and sequencing analysis revealed that her mother had R101Q, but the peak of the mutated base A is obviously lower than that of the normal G, indicating that only some cells of her mother contained the mutation (Figure 3), suggesting that the mother(II-7) is somatic mosaic for the mutation. The proband (III-5) and her sisters (III-6) had the clinical features of DS, but the phenotype of the mother was FS plus (FS+). Therefore, this is a generalized (genetic) epilepsy with FS plus (GEFS+) family, the phenotype of which include both FS+ and DS. Her father (II-8) had no seizure history and no mutation.
Parental origin of de novo mutations. Through AS-PCR and direct sequencing from DNA sample of the affected probands, we determined the parent origin of 12 de novo mutations (Figure 4). Parentage testing has confirmed the family relationship of the 12 probands and their parents. Ten mutations were paternal in origin (I91T, R101W, c.1662G>A, W768X, W952G, T1210K, V1390M, IVS10+3A>G, IVS15+1G>T, IVS18+2T>A) and two mutations were maternal in origin (F90S and IVS14+3A>T). The frequency of male germline transmission of the mutations is 83.3% (10 of 12).
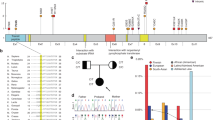
Schematic diagram of allele-specific PCR (AS-PCR). (A) SNPs 60096: C/G in the proband and his parents. The proband was heterozygous C/G: G was inherited from his father and C from his mother. M, mutant allele; W, wild-type allele. (B) Primers used to amplify the paternal and maternal allele of the proband. M, mutant allele; W, wild-type allele. (C) Sequencing result of the case number 27. (B) Source of SCN1A mutation IVS18+2T>A. Black arrows indicate the positions of the mutation in the proband. (a) Normal PCR. (b) Mutation was on the paternal allele. (c) The wild-type allele was from his mother.
Discussion
DS is a refractory epileptic syndrome characterized by the occurrence of FS before 1 year of age. This epileptic syndrome is diagnosed at 2–4 years of age after the progression of distinctive clinical features, thus it is often misdiagnosed as benign FS in its early stages. When children manifest repeated and prolonged FS triggered by fever, the possibility of DS should be considered.3 It has been reported that carbamazepine, oxcarbazepine and lamotrigine were ineffective to control seizures, and even aggravated seizure attacks in some DS patients.12, 13 Therefore, patients with this disease should avoid these drugs. In addition, valproic acid, topiramate, clonazepam and levetiracetam were effective in some patients.12, 13 SCN1A mutation analysis is helpful for DS patients in early diagnosis, selection of antiepileptic drugs, predicting the prognosis and genetic counseling. It should be considered as a routine investigation in suspected patients.
In the study presented here, the SCN1A mutation rate in DS was 77.8% (49 of 63), which is consistent with the international studies.7, 8, 14, 15 In the 49 mutations, only two mutations were recurrent in two unrelated cases. In all, there were 47 different mutations, of which 30 were novel to our best knowledge (Table 1).8, 16, 17 We found that SCN1A mutations in DS were individual and ‘hot spot’ mutations were still not identified. Using PCR-sequencing, we found 47 mutations, whereas by MLPA analysis one eight-exon duplication and one two-exon deletion were identified in two unrelated patients, respectively. SCN1A rearrangements of large fragments were responsible about 10% patients without mutations by PCR-sequencing, in which deletions were more frequent than duplications.18, 19 MLPA analysis was necessary for PCR-sequencing-negative patients, for about 12.5% (2 of 16) were abnormal. There were still 14 DS patients with no SCN1A mutations in our study, other genes such as SCN9A and PCDH19 may be involved in the pathogenesis of DS.19, 20 Our next step is to carry out SCN9A and PCDH19 mutation analysis in these patients.
There were various types of SCN1A mutations including missense, nonsense, splice-site and frame-shift mutations caused by base substitutions, deletions or insertions. These mutations were scattered in 26 exons of SCN1A, with mutations in exon 26 being the most prevalent. The mutagenesis analysis showed that the majority of base substitutions were caused by deamination and the second most common mutational mechanism being oxidation. About half of small deletion mutations were due to depurination, while the insertion mutations were more likely to be occurred in highly repetitive area. The causes of mutations should be further studied to better understand this severe disease.
The truncation mutations function as dominant negative, whereas the remaining normal genes show haploid insufficiency in the channel protein level.14 In addition to nonsense, frame-shift and splice-site mutations, large duplications or deletions of more than one exon can cause abnormal protein expression, thus truncating the sodium channel. In all, there were 30 truncation mutations accounting for 61.2% (30 of 49) in the total mutations. Functional studies have demonstrated that almost all SCN1A truncation mutations associated with DS markedly reduced inward sodium currents and cause loss of function of SCN1A.21 In addition, missense mutations located in the important functional areas (S4, S5, S6, S5–S6 linker) accounted for about half (8 of 19, 42.1%) of the total missense mutations. It was shown that mutations in the functional regions (S4–S6) in DS patients occurred more frequently than did those in GEFS+.22, 23 The other 11 missense mutations were either located in the highly conserved residues or these amino-acid substitutions changed the charged state of the residues. These characteristics of SCN1A mutations can partially explain the severity, refractory to antiepileptic drugs and poor prognosis features of DS.14, 23
Of the 49 patients with SCN1A mutations, DNA was available from 41 sets of parents. The mutations of 40 patients were initially identified to be de novo accounting for 97.6% (40 of 41), whereas one patient’s mutation was inherited from her mosaic mother. However, no familial mutations were identified. The majority of SCN1A mutations were de novo, consistent with SCN1A mutations having a significant role in the pathogenesis of DS. Patient number 5 and her sister both carry the mutation R101Q and both have DS. Their mutation was inherited from their mosaic mother, whose phenotype was FS+. As not all cells of their mother had the mutation R101Q, this most likely explains the reason why she has a less-severe phenotype than her two daughters. It was speculated that the expression level of mutant sodium channel in the neurons have not yet reached a ‘threshold’ of DS in their mother.24 Our study provides more evidence of parental mosaicisms in some DS patients.24, 25, 26 The possibility of mosaic mutations must be taken into account for genetic counseling.
In the majority of DS patients with SCN1A mutations, the somatic cells in their parents had no mutations, although PCR-sequencing may missed some somatic mosaic parents when the fraction of the mutant cells were trivial. The sequencing results suggested that all cells of the DS patients carried mutations. Therefore, these mutations were more likely to occur in the gamete period (male sperm or female ovum) before the formation of zygotes, otherwise they would be somatic mosaic. We use AS-PCR method to further detect the source of mutations.
In this research, it was not difficult to find suitable SNPs closer to the mutation site. Owing to SCN1A being located on the autosome 2q24.3, only when the SNPs of the child is heterozygous and the SNPs of one parent is homozygous, or the SNPs of both parents were different homozygous, we can determine the source of SNP and perform further AS-PCR to reveal the source of mutation. In 40 de novo mutations reported here, only 12 mutations were analyzed, of which 83.3% (10 of 12) were paternal. The results indicated that the majority of mutations were on the paternally derived chromosome, which is consistent with another recent study.27
Fathers of DS patients had more responsibility for the identified de novo mutations. We analyzed the reasons of the phenomenon as follows. It was known that there are obvious differences between the formation of male sperm and female ovum. The formation of sperm was more easily influenced by the external environment and mutagenic factors. In addition, DNA damages occurred more frequently in the replication period. The replication and meiotic division of spermatocytes were continuous and the sperm is generated throughout the man’s life, whereas all gene duplications of oocytes have been completed at birth.28 The oocytes only need to complete the second meiotic division to generate the ovum after birth. For women, there is only one ovum formation each month. Thus, most de novo mutations were paternal not only in DS, but also in other diseases, such as Apert syndrome and neurofibromatosis.28, 29, 30
Identification of the source of mutation provides an important theoretical basis for genetic counseling, rebirth guidance and prenatal diagnosis. In the genetic counseling, the parental mosaicism often caused recurrent transmission of SCN1A mutations to their offspring.24 Parents with no mutations in their somatic cells, who may be undetected gonadal mosaicism, also have more chances to bear another baby with the same mutations when they already have a child with SCN1A mutations. It means even for ‘de novo’ mutations, it was more possible that these mutations could reoccur in siblings.
In summary, SCN1A mutations were the major pathogenic gene for DS and there was no ‘hot spot’ mutation, which reinforces the mutational heterogeneity of SCN1A.7 MLPA analysis is an essential method for PCR-sequencing-negative DS patients. The vast majority of mutations were de novo, whereas rare mutations were inherited from their parents. Most de novo mutations were paternal. Recognition of SCN1A mutations in this severe epileptic syndrome assists in early diagnosis and selection of antiepileptic drugs.
Accession codes
References
Commission Classification and Terminology of the ILAE. Proposal for revised classification of epilepsies and epileptic syndromes. Epilepsia 30, 389–399 (1989).
Oguni, H., Hayashi, K., Osawa, M., Awaya, Y., Fukuyama, Y., Fukuma, G. et al. Severe myoclonic epilepsy in infancy: clinical analysis and relation to SCN1A mutations in a Japanese cohort. Adv. Neurol. 95, 103–117 (2004).
Caraballo, R. H. & Fejerman, N. Dravet syndrome: a study of 53 patients. Epilepsy Res. 70, 231–238 (2006).
Jansen, F. E., Sadleir, L. G., Harkin, L. A., Vadlamudi, L., McMahon, J. M., Mulley, J. C. et al. Severe myoclonic epilepsy of infancy (Dravet syndrome): recognition and diagnosis in adults. Neurology 67, 2224–2226 (2006).
Claes, L., Del-Favero, J., Ceulemans, B., Lagae, L., Van-Broeckhoven, C. & De-Jonghe, P. De novo mutations in the sodium-channel gene SCN1A cause severe myoclonic epilepsy of infancy. Am. J. Hum. Genet. 68, 1327–1332 (2001).
Claes, L., Ceulemans, B., Audenaert, D., Smets, K., Lofgren, A., Del-Favero, J. et al. De novo SCN1A mutations are a major cause of severe myoclonic epilepsy of infancy. Hum. Mutat. 21, 615–621 (2003).
Harkin, L.A., McMahon, J.M., Iona, X., Dibbens, L ., Pelekanos, J. T. & Zuberi, S.M. et al. The spectrum of SCN1A-related infantile epileptic encephalopathies. Brain 130 (3), 843–852 (2007).
Lossin, C. A catalog of SCN1A variants. Brain Dev. 31 (2), 114–130 (2009).
Baulac, S., Gourfinkel-An, I., Nabbout, R., Huberfeld, G., Serratosa, J., Leguern, E. et al. Fever, genes, and epilepsy. Lancet Neurol. 3 (7), 421–430 (2004).
Miller, S. A., Dykes, D. D. & Polesky, H. F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16 (3), 1215 (1988).
Ohmori, I., Ouchida, M., Ohtsuka, Y., Oka, E. & Shimizu, K. Significant correlation of the SCN1A mutations and severe myoclonic epilepsy in infancy. Biochem. Bioph. Res. Co. 295, 17–23 (2002).
Korff, C., Laux, L., Kelley, K., Goldstein, J., Koh, S. & Nordli, D. Dravet syndrome (severe myoclonic epilepsy in infancy): a retrospective study of 16 patients. J. Child. Neurol. 22, 186–194 (2007).
Striano, P., Coppola, A., Pezzella, M., Ciampa, C., Specchio, N., Ragona, F. et al. An open-label trial of levetiracetam in severe myoclonic epilepsy of infancy. Neurology 69 (3), 250–254 (2007).
Mulley, J., Scheffer, I., Petrou, S., Dibbens, L. M., Berkovic, S. F. & Harkin, L. A. SCN1A mutations and epilepsy. Hum. Mutat. 25, 535–542 (2005).
Fujiwara, T. Clinical spectrum of mutations in SCN1A gene: severe myoclonic epilepsy in infancy and related epilepsies. Epilepsy Res. 70S, 223–230 (2006).
Depienne, C., Trouillard, O., Saint-Martin, C., Gourfinkel-An, I., Bouteiller, D., Carpentier, W. et al. Spectrum of SCN1A gene mutations associated with Dravet syndrome: analysis of 333 patients. Med. Genet. 46 (3), 183–191 (2009).
Sun, H., Zhang, Y., Liang, J., Liu, X., Ma, X., Qin, J. et al. seven novel SCN1A mutations in Chinese patients with severe myoclonic epilepsy of infancy. Epilepsia. 49 (6), 1104–1107 (2008).
Mulley, J., Nelson, P., Guerrero, S., Dibbens, L., Iona, X., McMahon, J. M. et al. A new molecular mechanism for severe myoclonic epilepsy of infancy: exonic deletions in SCN1A. Neurology. 67, 1094–1095 (2006).
Singh, N. A., Pappas, C., Dahle, E. J., Claes, L. R., Pruess, T. H., De Jonghe, P. et al. A role of SCN9A in human epilepsies, as a cause of febrile seizures and as a potential modifier of Dravet syndrome. PLoS Genet. 5 (9), e1000649 (2009).
Depienne, C., Bouteiller, D., Keren, B., Cheuret, E., Poirier, K., Trouillard, O. et al. Sporadic infantile epileptic encephalopathy caused by mutations in PCDH19 resembles Dravet syndrome but mainly affects females. PLoS Genet. 5, e1000381 (2009).
Ohmori, I., Kahlig, K. M., Rhodes, T. H., Wang, D. W. & George, A. Nonfunctional SCN1A is common in severe myoclonic epilepsy of infancy. Epilepsia 47 (10), 1636–1642 (2006).
Kanai, K., Hirose, S., Oguni, H., Fukuma, G., Shirasaka, Y., Miyajima, T. et al. Effect of localization of missense mutations in SCN1A on epilepsy phenotype severity. Neurology 63, 329–334 (2004).
Ceulemans, B., Claes, L. & Lagae, L. G. Clinical correlations of mutations in the SCN1A gene: from febrile seizures to severe myoclonic epilepsy in infancy. Pediatr. Neurol. 30, 236–243 (2004).
Depienne, C., Arzimanoglou, A., Trouillard, O., Fedirko, E., Baulac, S., Saint-Martin, C. et al. Parental mosaicism can cause recurrent transmission of SCN1A mutations associated with severe myoclonic epilepsy of infancy. Hum. Mutat. 27 (4), 389 (2006).
Gennaro, E., Santorelli, F. M., Bertini, E., Buti, D., Gaggero, R., Gobbi, G. et al. Somatic and germline mosaicisms in severe myoclonic epilepsy of infancy. Biochem. Bioph. Res. Co. 341, 489–493 (2006).
Marini, C., Mei, D., Cross, H. & Guerrini, R. Mosaic SCN1A mutation in familial severe myoclonic epilepsy of infancy. Epilepsia. 47 (10), 1737–1740 (2006).
Heron, S. E., Scheffer, I. E., Iona, X., Zuberi, S. M., Birch, R., McMahon, J. M. et al. De novo SCN1A mutations in Dravet syndrome and related epileptic encephalopathies are largely of paternal origin. J. Med. Genet. 47 (2), 137–141 (2010).
Crow, J. F. The origins, patterns and implications of human spontaneous mutation. Nat. Rev. Genet. 1 (1), 40–47 (2000).
Moloney, D. M., Slaney, S. F., Oldridge, M., Wall, S. A., Sahlin, P., Stenman, G. et al. Exclusive paternal origin of new mutations in Apert syndrome. Nat. Genet. 13 (1), 48–53 (1996).
Jadayel, D., Fain, P., Upadhyaya, M., Ponder, M. A., Huson, S. M., Carey, J. et al. Parental origin of new mutations in von Recklinghausen neurofibromatosis. Nature 343, 558–559 (1990).
Acknowledgements
We are very grateful to all members of the families who participated in our study. We thank Dr Leanne M Dibbens (Department of Genetic Medicine, Women's and Children's Hospital, North Adelaide, South Australia, Australia) for revising the paper. This research was supported by grants from the Beijing Natural Science Foundation of China (No. 7072083).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Sun, H., Zhang, Y., Liu, X. et al. Analysis of SCN1A mutation and parental origin in patients with Dravet syndrome. J Hum Genet 55, 421–427 (2010). https://doi.org/10.1038/jhg.2010.39
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2010.39
Keywords
This article is cited by
-
Neurophysiology of the Flickering Light Perception
Neuroscience and Behavioral Physiology (2023)
-
Parental mosaicism in another case of Dravet syndrome caused by a novel SCN1A deletion: a case report
Journal of Medical Case Reports (2016)
-
The SCN1A gene variants and epileptic encephalopathies
Journal of Human Genetics (2013)