Abstract
An association study is a popular study design to identify susceptibility genes for common complex diseases. In such a study, the presence of inappropriate samples, such as those derived from close relatives or showing DNA contamination, causes an inflation of type I error or a decrease of power. Here we propose an identity-by-state (IBS)-based detection method of inappropriate samples taking linkage disequilibrium (LD) into consideration. The test statistics is the mean of the proportion of alleles that are shared identical by state at each single nucleotide polymorphism (SNP) between each sample pair in an association study. A covariance of the number of shared alleles between two SNPs is introduced to consider LD. We show that type I error and power are estimated accurately in computer-simulated data, and that if the number of SNPs analyzed is small, the performance of detection of inappropriate samples is superior to the previous method in simulated LD. An application to real association study data showed that accuracy in estimating the distribution of test statistics improved if LD was considered. Sample pairs considered to be siblings were detected. These results suggested that an LD-considered IBS-based detection method is useful in identifying inappropriate samples in an association study.
Similar content being viewed by others
Introduction
An association study is a popular study design to identify susceptibility genes for common complex diseases.1 Under the common disease-common variant (CD-CV) hypothesis,2 the power of an association study is generally higher than a linkage study for identification of disease susceptibility genes. Most association studies search for genetic markers that are related to a disease by comparing the frequency between the case (disease) and control (non-disease) populations. A disease-susceptibility gene may then be identified in the region of linkage disequilibrium (LD) corresponding to an associated genetic marker. Recently, biallelic single nucleotide polymorphisms (SNPs) are widely used as genetic markers.
A number of biases can be introduced in case–control association studies, making it very important to deal with them appropriately because they cause a significant inflation of type I error or a decrease of power. Quality control (QC), a series of operations to detect and remove biases, includes such possible causes as population stratification, sample contamination and cryptic relatedness.1, 3 Sample contamination can occur when samples of different individual origin are mixed by error in the experimental process of, for example, DNA extraction or SNP typing. Cryptic relatedness is observed when some close relatives are enrolled in a study by chance without the knowledge of investigators, which can cause an inflation of type I error.3
For general detection of related samples, a likelihood ratio test based on posterior probability of genotype under certain relationships was proposed.4 In the case of a family-based study, an identity-by-state (IBS)-based method5, 6 for a detection of errors in a sib-pair relationship was proposed, with the method using the summation of the IBS for a pair of sibs. Conversely, an identity-by-descent (IBD)-based method (PLINK7) was proposed. PLINK (http://pngu.mgh.harvard.edu/purcell/plink/) estimates genome-wide IBD-sharing coefficients between unrelated samples from genome-wide data. This metrics is useful for QC by diagnosing pedigree errors, undetected relationships, and sample swap, duplication and contamination events. It calculates π̂ (the proportion of alleles shared IBD) for each sample pairs, and contamination events are considered as outliers of π̂. In these previous studies, however, SNPs were assumed to be mutually independent, and LD was not taken into consideration. However, in many association studies, the LD between marker SNPs cannot be neglected.
Here we propose an IBS-based detection method to detect inappropriate samples (for example, contamination, close relatives) in an association study, which relies on SNP markers with or without LD. We evaluated a type I error and the power of the proposed method and estimated the number of SNPs required to detect inappropriate samples for marker SNPs in either LD or linkage equilibrium (LE). The proposed method was compared with the previous method by simulation. Finally, an application of the proposed method to an example of real data in a genome-wide association study suggested the practical relevance of our discussion.
Materials and methods
Test statistics
For K SNPs, the proposed test statistics is calculated as a mean proportion of alleles that are shared IBS over all SNPs. Let i and j denote the samples in an association study, and let Tk(i,j)ε{0, 0.5, 1} be the proportion of shared alleles at SNP k for sample (i, j) as follows:

Then we define the test statistics as follows:

It is assumed that almost all samples are not inappropriate (that is, no contamination or relatedness), so that the distribution of Yi,j under the null hypothesis H0 ((i,j) are independent) can be approximated by the normal distribution with mean μi,j and variance σ2i,j. No fixed effect for μi,j, σ2i,j was observed from real data, so that μi,j, σ2i,j are assumed to be independent of (i,j). Then μi,j≈μ, σi,j2≈σ2 are given by

We then take LD into account as covariance Cov between SNP k1 and SNP k2, and denote the range of LD by w, {w∣0<w<K}. In this study, we assume unrelated individuals, parent–child, siblings and contamination as the four types of sample pair relationships, denoted as R=1, 2, 3 and 4, respectively.
Parameter estimation and detection method
To estimate the E(Y) and V(Y), we note that E(Tk∣R=1), E(Tk2∣R=1) and Cov are expressed as

respectively, where pk and qk are the allele frequencies for SNP k (pk+qk=1), and the joint probability is calculated with the estimated haplotype frequency8 for SNP k1 and SNP k2 (see Mathematical details in Supplementary Information).
Tk of close relatives is higher than that of unrelated individuals, E(Tk∣R=3)>E(Tk∣R=2)>E(Tk∣R=1), and the genotypes of the contaminated samples should appear the same for each, so that inappropriate samples are detected as outliers (Y) of the distribution Y under the null hypothesis H0. In this method, the sample pairs whose Y is more than the threshold s={E(Y∣R=1)+E(Y∣R=2)}/2 are considered as outlier pairs.
Simulation study
We conducted two types of simulation. First, we evaluated the type I error and the power of the proposed method. Second, we compared the proposed method with the previous method in terms of the performance of type I error and power.
Simulation 1
We set the following conditions in the case of LE:
Condition 1: the number of samples (sum of case and control samples) is N=200, 600, 1000 and the number of SNPs is K=100, 200, 400, 800 and 1000.
Condition 2: under a null hypothesis H0 the allele frequency of each SNP has independent uniform distribution; the genotype frequency follows the Hardy-Weinberg equilibrium.
Condition 3: under an alternative hypothesis H1 there are three inappropriate sample pairs (parent–child, sibling and contaminated sample pairs, (Y∣R≠1)) in the simulation data.
Condition 4: type I error is estimated as u=#{Y∣Y>s,R=1}/(N(N−1)/2), which is the proportion of independent sample pairs (R=1) that are misjudged as inappropriate sample pairs. Power is estimated as v=#{Y∣Y>s,R=r},r=2,3,4, which is the number of inappropriate sample pairs that are detected correctly. In this simulation there is one inappropriate sample pair for each type (parent–child, sibling and contaminated sample pairs), so v=0 (not detected) or v=1 (detected). The threshold is s={E(Y∣R=1)+E(Y∣R=2)}/2.
Condition 5: the genotype of parent–child and sibling samples is assigned by using the conditional probability under each relationship.4 Genotypes of contaminated samples are assigned by the following process: if the genotypes of two samples at an SNP are the same, the genotypes of the two samples are not changed. If the genotypes are different, the genotypes of the two samples are assigned stochastically either as heterozygous or unchanged (the probability of the heterozygous and unchanged status is the same, 0.5 for each) for both samples.
The procedure to conduct the simulation experiment was as follows:
- Step 1.:
-
Set K and N according to the simulation conditions.
- Step 2.:
-
Generate SNP genotype data (Y∣R=1) under null hypothesis H0. Under alternative hypothesis H1 three normal sample pairs are replaced by inappropriate pairs (Y∣R≠1) in the original data of null hypothesis.
- Step 3.:
-
Calculate the threshold s for each data (H0 and H1), and estimate the u and v, respectively.
- Step 4.:
-
Repeat first three steps 1000 times and calculate the mean of u and v.
Simulation 2
We set the following conditions in the case of LD:
Condition 1: the number of samples (sum of case and control samples) is N=200, 600, 1000 and the number of SNPs is K=100, 200, 1000, 3000 and 5000.
Condition 2: under null hypothesis H0 the allele frequency of each SNP and the LD coefficient between SNPs is calculated from reference 100-SNP data selected from the HapMap (http://www.hapmap.org/) JPT data. The reference SNP data is composed of 10 regions on the genome; each region has 10 SNPs that are in the neighborhood of each other. In this simulation, these regions are called strong LD regions (w=10; Supplementary Figure 1).
Conditions 3 and 4: same as those previously described for the LE case.
Condition 5: the genotype of parent–child and siblings samples is assigned by the following process: first, any two specimens are sampled from association study data. Next, in each strong LD region the haplotype data of parent–child or sibling sample are generated by combining the four haplotypes of two samples according to Mendel's law. Genotypes of contaminated samples are assigned by the same process as in the case of LE.
The procedure to conduct the simulation experiment was as follows:
Steps 1, 3 and 4 are the same as those for LE.
Step 2: generate haplotype data based on the allele frequency and the coefficient of reference data by using bivariate normal distribution in each strong LD region.9 The association study SNP data are generated by the combination of any two haplotypes (Y∣R=1) under a null hypothesis H0. Under an alternative hypothesis H1 three normal sample pairs are replaced by inappropriate pairs (Y∣R≠1) in the original data of the null hypothesis.
We set the following parameters for PLINK (version 1.05) to compare the performance of the proposed method and IBD estimation of PLINK, ‘--genome --genome-full --min 0 --max 1’.
Real data
The proposed method was applied to our real association study data on 1498 samples and 2665 SNPs, which are the second screening data for the JSNP genome scan for gastric cancer.10 After a routine QC in the typing laboratory removed samples that have many missing values and a high proportion of heterozygotes, the allele frequency showed a nearly uniform distribution and there is a weak LD overall (Figure 1).
Results
Simulation Study
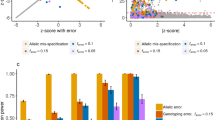
We evaluated the type I error and power (R=2, 3, 4) in the simulation data for SNP markers showing LE or LD (Tables 1 and 2). Type I error and power were calculated accurately by assuming the distribution of Y to be a normal distribution with mean E(Y) and variance V(Y) in both cases. In the case of LE, more than 800 SNPs were required to detect parent–child samples correctly (v̂=1) and to avoid exclusion of normal samples from the case–control data (ûN(N − 1)/2<1) (Table 1). Conversely, more than 3000 SNPs are required in the case of LD (Table 2). Because the correlation between the SNPs increases the variance V(Y), the type I error is inflated, the power decreases, and the necessary number of SNPs increases. Siblings or contaminated samples were also detected by the smaller number of SNPs (Tables 1 and 2). In comparing u0(w=0) and u10(w=10) in LD, type I error is calculated more accurately by taking LD into account (Table 2).
To compare the performance of the IBS-based method with the IBD-based method, we applied both methods to the simulation data with LD (K=200, N=200). In the IBD detection method, inappropriate samples were detected by the probability P(Z) of IBD state Z=z for the entire genome. The IBD-based method can detect parent–child samples by estimating P(Z=1), and can detect contaminated samples by estimating π̂=P(Z=1)/2+P(Z=2). As for parent–child samples, the IBS-based method in this simulation suppressed the false positive to a lower level than the IBD-based method. For contaminated samples, detection by the IBS-based method was more accurate than the IBD-based method (Figure 2). Sample contamination leads to too many heterozygote calls, which leads to fewer IBS 0 calls, which leads to an overestimated IBD and increased π̂. However, in the contamination process simulated in this study, the genotypes of the contaminated samples were stochastically assigned as heterozygote and the IBD estimates did not increase as much, so the performance of the IBD-based method was low.
Although the number of SNPs is insufficient to detect inappropriate samples accurately according to Table 2, we focus in this simulation on the association study in which the number of SNPs is less than 1000. Moreover, we confirm that there is no difference in the performance between the two methods in the case of 1000 SNPs, and that both methods detect inappropriate samples accurately (data not shown).
Real data analysis
We applied the IBS-based method to the real association study data while changing the number of SNPs (K=200, 600, 1000 and 2665). This real data had a weak LD overall (Figure 1). It was possible to approximate the distribution of Y by a normal distribution, and there was little difference between w=10 and w=100 (Figure 3). In the case of a weak LD, the estimation accuracy of Y could be improved by considering LD. The number of detected sample pairs was estimated accurately by an upper probability of normal distribution (Table 3). The detected two sample pairs were rechecked by clinical-side investigators, and a sibling relationship was in fact strongly suggested.
Discussion
In an association study, a series of QC is essential to maintain research quality. In this study, we focused on the detection of inappropriate samples. Previously, the IBS-based detection methods were proposed in family-based studies.5, 6 However, these methods did not consider LD among genetic markers and thus cannot be applied to association study data with LD. Our new IBS-based detection method can consider LD by using the covariance of Y, and the type I error and the power of the proposed method were able to be evaluated accurately by a simulation study. In a typical association study with only a few inappropriate samples, it is necessary to evaluate type I error correctly to avoid inadvertent exclusion of appropriate samples. In the simulation data, the proposed method detected inappropriate samples correctly and more accurately than did the IBD-based method.
In our simulation study the number of false positive drastically decreases when more than 1000 SNPs are analyzed (Table 2), and the PLINK website also reports that a large number of SNPs (1000 independent SNPs at a minimum) is required to calculate genome-wide IBD given IBS information. Taken together, this means that more than 1000 SNPs are required to detect inappropriate samples accurately. However, in some candidate gene approaches the target genes have been defined already and the number of typing SNPs on these genes is less than 1000 SNPs. In such a case we recommend the proposed method.
In the proposed method, we set the threshold s={E(Y∣R=1)+E(Y∣R=2)}/2. Setting the optimal threshold by using Bayes factor6 is necessary on the assumption that the distribution of Y is a mixed normal distribution of unrelated (R=1) and inappropriate samples (parent–child (R=2) and siblings (R=3) and so on). However, as inappropriate samples are generally infrequent, it is difficult to estimate the mixed rate and parameter of the inappropriate samples’ distribution. Thus we simply adopt the threshold defined by s={E(Y∣R=1)+E(Y∣R=2)}/2. There is room for study on how to decide the threshold.
In the proposed method, we assumed a virtual strong LD region as consecutive SNPs, and the covariance Cov is calculated within this region. Because the LD pattern is variable across the genome, it is reasonable to consider the covariance according to the position-dependent LD width. However, the results of the real data suggested that it is acceptable to regard the strong LD region as a region that consists of a number of consecutive SNPs.
In the application to real data, we excluded beforehand the samples that have many missing SNPs or a high proportion of heterozygous SNPs, because doing so is part of the routine QC process in our typing laboratory. In fact, we found that the inclusion of these samples inflates the variance of Y, which in turn overestimates the type I error. In our current QC procedure, we do not consider LD in the detection and exclusion of samples with an inappropriately high proportion of heterozygosity. A method that considers LD in a similar manner to the proposed one can be applied to the detection of a sample with a high proportion of heterozygosity by using Tk=1(the genotype is heterozygous at SNP k), Tk=0 (the genotype is homozygous at SNP k). Please note that a nonreciprocal, unidirectional contamination, in which sample B is contaminated with sample A, while sample A remains untouched, can be detected by the abnormally high proportion of heterozygosity of the contaminated sample B.
Recently, the introduction of powerful array-based SNP typing platforms has made a genome-wide association study a popular strategy for identifying disease-associated genes, and genotype data on 100 000–1 000 000 SNPs are increasingly available. In a genome-wide association study, inappropriate samples can be efficiently detected, because it is possible to select several hundreds of SNPs for quality control purposes (QC-SNP). It is necessary to select QC-SNPs that are in LE with each other and whose allele frequencies are around 0.5; such SNPs can most efficiently distinguish inappropriate samples from normal ones. On the other hand, when a few candidate genes or a genomic region of interest are already known or selected, and a high-density SNP typing is desired on these genes, it is necessary to consider LD by the proposed method.
In this study, we proposed a detection method for inappropriate sample pairs in a case–control association study. When we applied the proposed method to real association study data, two sample pairs were detected as siblings. Once inappropriate samples are strongly suspected, we usually take the following action: when contamination is detected, we exclude all relevant samples from the case–control data. If a related sample pair is detected, we usually keep only one subject from the pair by a combination of the following two criteria: (1) a case subject is selected if the pair includes both case and control subjects, because case subjects are more limited in availability than controls in many association studies; (2) the overall typing data quality of the samples, particularly the SNP call rate (number of the successfully genotyped SNPs for each sample). However, if the number of inappropriate samples is substantial, the decision whether to include them may require the consideration of a trade-off between an inflation of type I error and decreased power of the test. In this case, we may need a future study on sensitivity analysis to evaluate the trade-off.
References
Balding, D. J., Bishop, M & Cannings, C. Handbooks of Statistical Genetics 3rd edn. (Wiley, Chichester, 2007).
Wright, A. F. & Hastie, N. D. Complex genetic diseases: controversy over the Croesus code. Genome Biol. 2, COMMENT2007.1–COMMENT2007.8. (2001).
Voight, B. F. & Pritchard, J. K. Confounding from cryptic relatedness in case–control association studies. PLoS Genet. 1, e32 (2005).
Wenk, R. E., Traver, M. & Chiafari, F. A. Determination of sibship in any two persons. Transfusion 36, 259–262 (1996).
Ehm, M. G. & Wagner, M. A test statistic to detect errors in sib-pair relationship. Am. J. Hum. Genet. 62, 181–188 (1998).
Zhang, B. & Betensky, R. A. Methods to classify familial relationships in the presence of laboratory errors, without parental data. Hum. Genet. 119, 642–648 (2006).
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A. R., Bender, D. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Excoffier, L. & Slatkin, M. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol. Biol. Evol. 12, 921–927 (1995).
Giovanni, M. HapSim: a simulation tool for generating haplotype data with pre-specified allele frequencies and LD coefficients. Bioinformatics 21, 4309–4311 (2005).
Study Group of Millennium Genome Project for Cancer, Sakamoto, H. Yoshimura, K. Saeki, N. Katai, H., Shimoda, T. et al. Genetic variation in PSCA is associated with susceptibility to diffuse-type gastric cancer. Nat. Genet. 40, 730–740 (2008).
Acknowledgements
We thank K Yoshimura, Shumpei Ohnami, Sumiko Ohnami, N Saeki, A Kuchiba, H Totsuka, A Saito, S Chiku at the National Cancer Center for their valuable advice. This study was supported by the Program for the Promotion of Fundamental Studies in Health Sciences from the National Institute of Biomedical Innovation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Andoh, M., Sato, Y., Sakamoto, H. et al. Detection of inappropriate samples in association studies by an IBS-based method considering linkage disequilibrium between genetic markers. J Hum Genet 55, 436–440 (2010). https://doi.org/10.1038/jhg.2010.43
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2010.43