Abstract
Serotonin (5-hydroxytryptamine (5-HT)) transporter (SLC6A4) is known to influence mood, emotion, cognition and efficacy of antidepressants, particularly that of selective serotonin reuptake inhibitors. Atypical antipsychotics exert their effects partially through serotinergic systems, and hence, variation in 5-HT uptake may affect antipsychotic action mediated through the serotinergic system. Therefore, investigating the role of SLC6A4 as a risk factor for developing schizophrenia and treatment response had been a point of concern for many investigators, but with variable outcome. In this study, we examined the genetic roles of five polymorphisms of SLC6A4, including those of the widely studied 44 base pair variable number of tandem repeat (VNTR) in the promoter region of SLC6A4 (the serotonin transporter gene-linked polymorphic region: 5HTTLPR) and a VNTR polymorphism (STin2) in the second intron, in schizophrenia and its influence on the severity of symptoms in a South Indian population from Kerala, comprising 586 individuals. We detected significant allelic and genotypic associations with rs2066713 (both allelic and genotypic P-value <0.001), 5HTTLPR (allelic P-value=0.008 and genotypic P-value=0.03) and STin2 polymorphisms (allelic P-value=0.001 and genotypic P-value=0.002). A haplotype linking these three risk alleles, 5HTTLPR/S-rs2066713/C-STin2/12-repeat (P-value=0.0059), was also significantly associated with disease in our population. Patients with STin2 12-repeat homozygotes showed a greater severity of blunted effect symptom. These results suggest a strong role of SLC6A4 in schizophrenia, possibly with a specific behavioral endophenotype in a South Indian population.
Similar content being viewed by others
Introduction
Schizophrenia is a common and complex psychiatric disorder, with proven genetic causes. Research to unravel the genetic basis of schizophrenia has revealed several molecules, including those involved in dopamine and serotonin neurotransmitter systems for the past few decades. Serotonin (5-hydroxytryptamine (5-HT)) hypothesis of schizophrenia has been reanalyzed with the effectiveness of clozapine, the best known atypical antipsychotic drug,1 as an antagonist for several 5-HT receptors.2, 3 Moreover, 5-HT has been shown to regulate the development of the central nervous system. As the 5-HT transporter (5-HTT) regulates the 5-HT system,4 alterations in the function of this protein could be involved in the development of schizophrenia.5
The serotonin transporter (SLC6A4) gene spanning 37.8 kb is located on chromosome 17q11.2,6, 7 and has 14 exons encoding a protein of 630 amino acids.8 SLC6A4 has received attention as a candidate in schizophrenia and mood disorders, owing to its influence on mood, emotion, cognition and also because 5-HTT is the principal site of action for some antidepressants, particularly for the selective serotonin reuptake inhibitors.9 Serotonin transporter is responsible for removing 5-HT from the synaptic cleft.10 5-HTT removes 5-HT from the synaptic cleft and decides the extent and period of postsynaptic receptor-mediated signaling, having a crucial function in the fine-tuning of 5-HT neurotransmission.11, 12, 13 The synaptic clearance of 5-HT is determined by the number of 5-HTTs expressed at the presynaptic surface.14 Release and active reuptake of serotonin determine the extent and duration of chemical signaling and thus, exert a key effect in the spatio-temporal fine-tuning of serotonergic neurotransmission.
A number of studies have screened SLC6A4 polymorphisms and examined their associations with several psychiatric diseases, including schizophrenia. Of these, a 44 base pair insertion/deletion variable number of tandem repeat (VNTR) located in the promoter region of SLC6A4 (the serotonin transporter gene-linked polymorphic region: 5HTTLPR)6, 15 and a VNTR polymorphism (STin2) in the second intron16 have been studied extensively. Atypical antipsychotics exert their effects partially through serotoninergic systems, and hence, variation in 5-HT uptake may also affect antipsychotic action, which is mediated through the serotonergic system.4
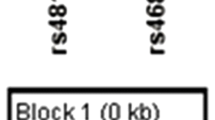
In this study, we investigated the role of five polymorphisms from SLC6A4 in schizophrenia and their influence on the severity of symptoms in a South Indian population from Kerala. Single-nucleotide polymorphisms (SNPs) in the serotonin transporter gene have been selected on the basis of their functional significance or on evidence of being a tag SNP. SLC6A4 has two blocks of strong linkage disequilibrium (LD), in which 5HTTLPR and STin2 lie in different LD blocks, and it has been suggested that this difference could be the reason for inconsistency in earlier associations with SLC6A4 polymorphisms.17 5HTTLPR and STin2 polymorphisms are reported to alter expression profile. rs1042173 is located in a putative miRNA-binding site in the 3′-end of the gene and rs2066713 and rs11080121 are tag SNPs located in the 5′-and 3′-regions, respectively.
Materials and methods
Subjects included 243 drug-naive schizophrenic patients (82 men and 161 women), with a mean age of 33±11 years from Kerala, South India. All patients were diagnosed on the basis of DSM-IV (Diagnostic and Statistical Manual of Mental Disorders) criteria.18 Baseline rating of these drug-naive patients on the basis of the BPRS-E (Extended Brief Psychiatric Rating Scale, 24 item, score 1–7) was carried out at the time of first consultation.19 Control samples included 243 ethnically and age-matched (31.7±6.92) normal volunteers from Kerala, India. Control individuals had no family history of schizophrenia and had never taken neuroleptics in their lifetime.
Genotyping
Peripheral blood was collected from all individuals in EDTA (ethylenediaminetetraacetic acid)-coated vials. The study was approved by the Institutional ethics committee for biomedical subjects, which met the Indian Council for Medical Research guidelines. Genomic DNA was isolated from lymphocytes according to Sambrook and Russell.20 Selected polymorphisms in SLC6A4 were genotyped using the primer sets described in Table 1. We analyzed five polymorphisms, of which 5HTTLPR, rs2066713 and STin2 were located around the 5′-region, whereas rs11080121 and rs1042173 were located near the 3′-end (Figure 1). SNPs found in SLC6A4 have been reported to fall in three blocks of strong LD, of which functional polymorphisms 5HTTLPR and STin2 lie in different LD blocks in the 5′-end, whereas rs1042173 falls in the third block and is located in the 3′-end. rs2066713 and rs11080121 are reported as tag SNPs in the 5′- and 3′-regions, respectively.
For 5HTTLPR genotyping, DNA was amplified using Pwo superyield DNA polymerase (Roche Applied Science, Mannheim, Germany) 1.25 U, 1 × GC-rich resolution solution buffer, 5 mM dNTP (C7-dGTP:dGTP=1:1) and 5 mM of forward and reverse primers (Table 1). After initial denaturation at 98 °C for 5 min, 35 cycles were carried out at 98 °C for 45 s, at 68 °C for 1 min, followed by a final step of elongation at 68 °C for 10 min. The product lengths were 221 bp (short allele) and 265 bp (long allele). PCR products were resolved on 3% agarose gel stained with ethidium bromide.
For STin2.VNTR typing, DNA was amplified using platinum Taq polymerase (Invitrogen, Carlsbad, CA, USA) 1.5 U, 1 × buffer, 5 mM dNTP and 5 mM of forward and reverse primers (Table 1). Touchdown protocol was used as follows: after an initial denaturation at 95 °C for 3 min, five touchdown cycles were carried out at an annealing temperature from 65 °C for 1 min to 63 °C, with a decrement of 0.4 °C per cycle and an extension of 72 °C for 1 min. The remaining 35 cycles were carried out with an annealing temperature of 63 °C for 1 min, followed by a final step of elongation at 72 °C for 10 min. The PCR products were run on 2% agarose gel stained with ethidium bromide. The 12-repeat variant (STin2.12) showed the fragment of 299 bp, and the 10-repeat variant (STin2.10) showed the fragment of 265 bp.
Single-nucleotide polymorphisms rs2066713, rs11080121 and rs1042173 were genotyped by sequencing. A PCR product of 1 μl was used for sequencing reaction (ABI PRISM BigDye Terminator v3.1 cycle sequencing kit) according to the manufacturer's instructions, and was analyzed using the ABI PRISM 3730 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Statistical analyses
Allelic and genotypic distributions were tested for association using Fisher's exact test. Deviations of genotype distributions from the Hardy–Weinberg equilibrium (HWE) were evaluated by the χ2 test (degrees of freedom (d.f.)=1). Haplotypic association of samples was carried out using the COCAPHASE program in the UNPHASED v3.011 package (http://www.mrc-bsu.cam.ac.uk/personal/frank/software/unphased/) with 10 000 permutations.21 To estimate LD between pairs of loci in the patient and control populations, standardized disequilibrium coefficient (D′) and squared correlation coefficient (r2) were calculated using Haploview 4.1 (http://www.broad.mit.edu/mpg/haploview/).22 LD blocks were defined in accordance with Gabriel's criteria.23 Statistical power for detecting association was calculated using the Genetic Power Calculator (http://pngu.mgh.harvard.edu/~purcell/gpc/)24 under the following parameter assumption: genetic relative risk=1.5, prevalence of disease=0.01, risk allele frequency=0.3, α=0.05 and a multiplicative model of inheritance. Kruskal–Wallis ANOVA (analysis of variance) was used to investigate the contribution of SLC6A4 genotypes to the symptoms of patients.
Results
In both cases and controls, all five polymorphisms were in HWE, with the exception of rs2066713 in the patient group. The allelic and genotypic frequencies of SLC6A4 polymorphisms are listed in Table 2. Strong allelic and genotypic associations with schizophrenia were observed with 5HTTLPR (P=0.008 and P=0.03, respectively), rs2066713 (both P-values <0.001) and STin2 (P=0.001 and P=0.002, respectively). All P-values, except the 5HTTLPR genotypic P-value, remained significant after Bonferroni's correction. We also observed higher homozygous genotypic frequencies of the short allele of 5HTTLPR, the C allele of rs2066713 and the 12-repeat of Stin2 polymorphisms in the patient group. The SNPs located near the 3′-end of the gene, rs11080121 and rs1042173 were not associated with schizophrenia. We further evaluated the LD between markers using COCAPHASE. Substantial LD blocks were observed between rs2066713 and Stin2 and also between rs11080121 and rs1042173 in cases and controls, although the former two SNPs were not defined as an LD block (Figure 2). 5HTTLPR was in moderate LD with rs2066713 (D′=0.54 in the case group and D′=0.50 in the control group) and STin2 (D′=0.58 in the case group and D′=0.62 in the control group) polymorphisms.
On the basis of the fact that three 5′ polymorphisms showed significant associations with schizophrenia and that they are in moderate or substantial LD to each other, we performed haplotype analyses against this genomic region using COCAPHASE. The three-locus haplotype, 5HTTLPR/S-rs2066713/C-STin2/12-repeat (S-C-12), was significantly higher in patients than in controls (P=0.0009; after 10 000 permutations, P=0.0059; global P=0.0039) (Table 3). We further examined the contribution of SLC6A4 genotypes to the symptoms of patients. Our study showed a significant effect of the STin2 12-repeat homozygote on the higher intensity of blunted effect (Kruskal–Wallis' statistic=6.78, d.f.=2, P=0.03). The power analyses disclosed that this study has a power of 36% to detect a genotype relative risk of 1.5 (α=0.05) under a multiplicative model.
Discussion
In this study, we detected significant associations of 5HTTLPR, rs2066713 and STin2 located near the 5′-region of SLC6A4 with schizophrenia in a South Indian population. The short allele of 5HTTLPR, the C allele of rs2066713 and the 12-repeat of STin2 were observed as risk alleles, and their homozygous genotypes were more frequent in the schizophrenia group than in the control group. A haplotype consisting of the S-C-12-repeat of three risk alleles was also associated with schizophrenia in our population. A large number of studies have been carried out to investigate the role of SLC6A4 polymorphisms in schizophrenia. A majority of these studies have concentrated on 5HTTLPR and STin2 polymorphisms. The transcriptional activity of the long variant of 5HTTLPR was 1.4–1.7 times greater than that of the short form of 5HTTLPR and could bind 30–40% more of a labeled marker.6 These observations suggest that the short allele of 5HTTLPR has reduced promoter activity, resulting in a decreased gene expression and low serotonin uptake.15 The short allele has been reported to have a modest role in anxiety-related personality trait25 and suicidal behavior.26
Within SLC6A4, STin2 has been reported to be the strongest candidate polymorphism showing susceptibility to schizophrenia.17, 27 The STin 2.12 allele has been reported to have increased vulnerability to schizophrenia.28, 29, 30, 31 These observations are consistent with current results. Functionally, the alleles of STin2 VNTR (STin2.10 and STin2.12) can act as transcriptional regulatory elements, with STin2.12 having a superior enhancer-like property within the developing rostral hindbrain, which may lead to aberrant serotonergic neuron development.32, 33 In spite of the evidence for functional effects of alleles at STin2 VNTR, it is not possible to determine whether this VNTR is causally associated with increased risk of schizophrenia. It has been suggested that individual polymorphisms have a weak influence when compared with the combined effect of 5-HTTLPR and STin2 VNTR polymorphisms on SLC6A4 expression.31 STin2 VNTR may also have an effect as a tissue-specific transcriptional regulator of SLC6A4,34 which may be critical in the presentation of individual symptoms. In our study, the STin2 12-repeat homozygote was associated with blunted effect. A blunted response may be relevant to decreased levels of serotonin availability, because the homozygous STin2 12-repeat could result in an increased serotonin uptake, therefore eliminating serotonin in the synaptic cleft efficiently.35 Our current data correlate well with published studies and meta-analyses of SLC6A4 in schizophrenia, in which the S allele of 5HTTLPR and the 12-repeat allele of STin2 VNTR have been associated with schizophrenia.17, 36 In a case–control association study, we have to pay attention to sample stratification. In this study, we selected the patients and controls from a genetically characterized Malayalam-speaking Dravidian community from Kerala.37
Ethnicity and heterogeneity within and between population groups could affect the haplotypic diversity and pattern of LD in study populations. Significant global variation in control subjects has been observed in the allelic frequencies of 5HTTLPR and STin2 VNTR polymorphisms (Supplementary Tables 1 and 2). The allelic frequencies in our South Indian populations are comparable with those in populations of European descent. Chinese, Japanese and Korean populations show an increased representation of the S allele of 5HTTLPR and the 12-repeat of STin2 polymorphisms, in comparison with our data and with those of European descent. It has been reported that LD between 5HTTLPR and STin2 varies from strong in native Americans to weak in Europeans and is absent in Chinese.38 Our data revealed an LD block structure similar to that of Europeans,17 displaying high D′ values between markers rs2066713 and STin2, and between rs11080121 and rs1042173.
In conclusion, the serotonin transporter gene has a potential susceptibility for schizophrenia in our Indian samples, although the exact mechanisms of its promoter and of other 5′-side variants for schizophrenia development remain to be elucidated.
References
Meltzer, H. Y. An overview of the mechanism of action of clozapine. J. Clin. Psychiatry 55 (Suppl B), 47–52 (1994).
Roth, B. L., Ciaranello, R. D. & Meltzer, H. Y. Binding of typical and atypical antipsychotic agents to transiently expressed 5-HT1C receptors. J. Pharmacol. Exp. Ther. 260, 1361–1365 (1992).
Watling, K. J., Beer, M. S., Stanton, J. A. & Newberry, N. R. Interaction of the atypical neuroleptic clozapine with 5-HT3 receptors in the cerebral cortex and superior cervical ganglion of the rat. Eur. J. Pharmacol. 182, 465–472 (1990).
Lesch, K. P. & Mossner, R. Genetically driven variation in serotonin uptake: is there a link to affective spectrum, neurodevelopmental, and neurodegenerative disorders? Biol. Psychiatry 44, 179–192 (1998).
Torres, G. E., Gainetdinov, R. R. & Caron, M. G. Plasma membrane monoamine transporters: structure, regulation and function. Nat. Rev. Neurosci. 4, 13–25 (2003).
Heils, A., Teufel, A., Petri, S., Stober, G., Riederer, P. & Bengel, D. et al. Allelic variation of human serotonin transporter gene expression. J. Neurochem. 66, 2621–2624 (1996).
Lesch, K. P., Balling, U., Gross, J., Strauss, K., Wolozin, B. L., Murphy, D. L. et al. Organization of the human serotonin transporter gene. J. Neural. Transm. Gen. Sect. 95, 157–162 (1994).
Ramamoorthy, S., Bauman, A. L., Moore, K. R., Han, H., Yang-Feng, T., Chang, A. S. et al. Antidepressant- and cocaine-sensitive human serotonin transporter: molecular cloning, expression, and chromosomal localization. Proc. Natl Acad. Sci. USA 90, 2542–2546 (1993).
Schloss, P. & Williams, D. C. The serotonin transporter: a primary target for antidepressant drugs. J. Psychopharmacol. 12, 115–121 (1998).
Lesch, K. P., Greenberg, B. D. & Higley, J. D. Molecular Genetics of Human Personality. Benjamin, J., Ebstein, R.P., and Belmaker, R.H. Washington, DC (2002).
Blakely, R. D., De Felice, L. J. & Hartzell, H. C. Molecular physiology of norepinephrine and serotonin transporters. J. Exp. Biol. 196, 263–281 (1994).
Uhl, G. R. & Johnson, P. S. Neurotransmitter transporters: three important gene families for neuronal function. J. Exp. Biol. 196, 229–236 (1994).
Lesch, K. P., Meyer, J., Glatz, K., Flugge, G., Hinney, A., Hebebrand, J. et al. The 5-HT transporter gene-linked polymorphic region (5-HTTLPR) in evolutionary perspective: alternative biallelic variation in rhesus monkeys. Rapid communication. J. Neural. Transm. 104, 1259–1266 (1997).
Beckman, M. L. & Quick, M. W. Neurotransmitter transporters: regulators of function and functional regulation. J. Membr. Biol. 164, 1–10 (1998).
Lesch, K. P., Bengel, D., Heils, A., Sabol, S. Z., Greenberg, B. D., Petri, S. et al. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science 274, 1527–1531 (1996).
Ogilvie, A. D., Battersby, S., Bubb, V. J., Fink, G., Harmar, A. J., Goodwim, G. M. et al. Polymorphism in serotonin transporter gene associated with susceptibility to major depression. Lancet 347, 731–733 (1996).
Fan, J. B. & Sklar, P. Meta-analysis reveals association between serotonin transporter gene STin2 VNTR polymorphism and schizophrenia. Mol. Psychiatry 10, 928–938, 891 (2005).
Diagnostic and statistical manual of mental disorders (DSM-IV) (American Psychiatric Association, Washington, DC, 1994).
Dingemans, P. M., Linszen, D. H., Lenior, M. E. & Smeets, R. M. Component structure of the Expanded Brief Psychiatric Rating Scale (BPRS-E). Psychopharmacology (Berl) 122, 263–267 (1995).
Sambrook, J. & Russell, D. W. Molecular Cloning: A Laboratory Manual (CSHL, New York, 2001).
Dudbridge, F. Pedigree disequilibrium tests for multilocus haplotypes. Genet. Epidemiol. 25, 115–121 (2003).
Barrett, J. C., Fry, B., Maller, J. & Daly, M. J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005).
Gabriel, S. B., Schaffner, S. F., Nguyen, H., Moore, J. M., Roy, J., Blumenstiel, B. et al. The structure of haplotype blocks in the human genome. Science 296, 2225–2229 (2002).
Purcell, S., Cherny, S. S. & Sham, P. C. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 19, 149–150 (2003).
Munafo, M. R., Freimer, N. B., Ng, W., Ophoff, R., Veijola, J., Miettunen, J. et al. 5-HTTLPR genotype and anxiety-related personality traits: a meta-analysis and new data. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 150B, 271–281 (2008).
Neves, F. S., Silveira, G., Romano-Silva, M. A., Malloy-Diniz, L., Ferreira, A. A., De Marco, L. et al. Is the 5-HTTLPR polymorphism associated with bipolar disorder or with suicidal behavior of bipolar disorder patients? Am. J. Med. Genet. B Neuropsychiatr. Genet. 147B, 114–116 (2008).
Allen, N. C., Bagade, S., McQueen, M. B., Ioannidis, J. P., Kavvoura, F. K., Khoury, M. J. et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat. Genet. 40, 827–834 (2008).
Collier, D. A., Arranz, M. J., Sham, P., Battersby, S., Vallada, H., Gill, P. et al. The serotonin transporter is a potential susceptibility factor for bipolar affective disorder. Neuroreport 7, 1675–1679 (1996).
Liu, W., Gu, N., Feng, G., Li, S., Bai, S., Zhang, J. et al. Tentative association of the serotonin transporter with schizophrenia and unipolar depression but not with bipolar disorder in Han Chinese. Pharmacogenetics 9, 491–495 (1999).
Kaiser, R., Tremblay, P. B., Schmider, J., Henneken, M., Dettling, M., Müller-Oerlinghausen, B. et al. Serotonin transporter polymorphisms: no association with response to antipsychotic treatment, but associations with the schizoparanoid and residual subtypes of schizophrenia. Mol. Psychiatry 6, 179–185 (2001).
Hranilovic, D., Stefulj, J., Schwab, S., Borrmann-Hassenbach, M., Albus, M., Jernej, B. et al. Serotonin transporter promoter and intron 2 polymorphisms: relationship between allelic variants and gene expression. Biol. Psychiatry 55, 1090–1094 (2004).
MacKenzie, A. & Quinn, J. A serotonin transporter gene intron 2 polymorphic region, correlated with affective disorders, has allele-dependent differential enhancer-like properties in the mouse embryo. Proc. Natl Acad. Sci. USA 96, 15251–15255 (1999).
Lovejoy, E. A., Scott, A. C., Fiskerstrand, C. E., Bubb, V. J. & Quinn, J. P. The serotonin transporter intronic VNTR enhancer correlated with a predisposition to affective disorders has distinct regulatory elements within the domain based on the primary DNA sequence of the repeat unit. Eur. J. Neurosci. 17, 417–420 (2003).
Fiskerstrand, C. E., Lovejoy, E. A. & Quinn, J. P. An intronic polymorphic domain often associated with susceptibility to affective disorders has allele dependent differential enhancer activity in embryonic stem cells. FEBS Lett. 458, 171–174 (1999).
Coutinho, A. M., Oliveira, G., Morgadinho, T., Fesel, C., Macedo, T. R., Bento, C. et al. Variants of the serotonin transporter gene (SLC6A4) significantly contribute to hyperserotonemia in autism. Mol. Psychiatry 9, 264–271 (2004).
Zaboli, G., Jonsson, E. G., Gizatullin, R., De Franciscis, A., Asberg, M. & Leopardi, R. Haplotype analysis confirms association of the serotonin transporter (5-HTT) gene with schizophrenia but not with major depression. Am. J. Med. Genet. B Neuropsychiatr. Genet. 147, 301–307 (2008).
Thomas, R., Nair, S. B. & Banerjee, M. HLA-B and HLA-C alleles and haplotypes in the Dravidian tribal populations of southern India. Tissue Antigens 64, 58–65 (2004).
Gelernter, J., Cubells, J. F., Kidd, J. R., Pakstis, A. J. & Kidd, K. K. Population studies of polymorphisms of the serotonin transporter protein gene. Am. J. Med. Genet. 88, 61–66 (1999).
Acknowledgements
NNV acknowledges the University Grants Commission (UGC), Government of India for providing junior research fellowship, and Riken Brain Science Institute (RIKEN BSI), Japan for summer training fellowship. We also acknowledge all the patients and their family members who cooperated in this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website (http://www.nature.com/jhg)
Supplementary information
Rights and permissions
About this article
Cite this article
Vijayan, N., Iwayama, Y., Koshy, L. et al. Evidence of association of serotonin transporter gene polymorphisms with schizophrenia in a South Indian population. J Hum Genet 54, 538–542 (2009). https://doi.org/10.1038/jhg.2009.76
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2009.76
Keywords
This article is cited by
-
Serotonin transporter functional polymorphisms potentially increase risk of schizophrenia separately and as a haplotype
Scientific Reports (2022)
-
A three-pronged analysis confirms the association of the serotoninergic system with attention deficit hyperactivity disorder
World Journal of Pediatrics (2022)
-
Attempts to replicate genetic associations with schizophrenia in a cohort from north India
npj Schizophrenia (2017)
-
Association and interaction analyses of 5-HT3 receptor and serotonin transporter genes with alcohol, cocaine, and nicotine dependence using the SAGE data
Human Genetics (2014)
-
Association between a genetic variant in the serotonin transporter gene (SLC6A4) and suicidal behavior in patients with schizophrenia
Behavioral and Brain Functions (2012)