Abstract
Duplications of one or more exons of the dystrophin gene are the second most common mutation in dystrophinopathies. Even though duplications are suggested to occur with greater complexity than thought earlier, they have been considered an intragenic event. Here, we report the insertion of a part of the IL1RAPL1 (interleukin-1 receptor accessory protein-like 1) gene into the duplication junction site. When the actual exon junction was examined in 15 duplication mutations in the dystrophin gene by analyzing dystrophin mRNA, one patient was found to have an unknown 621 bp insertion at the junction of duplication of exons from 56 to 62. Unexpectedly, the inserted sequence was found completely identical to sequences of exons 3–5 of the IL1RAPL1 gene that is nearly 100 kb distal from the dystrophin gene. Accordingly, the insertion of IL1RAPL1 exons 3–5 between dystrophin exons 62 and 56 was confirmed at the genomic sequence level. One junction between the IL1RAPL1 intron 5 and dystrophin intron 55 was localized within an Alu sequence. These results showed that a fragment of the IL1RAPL1 gene was inserted into the duplication junction of the dystrophin gene in the same direction as the dystrophin gene. This suggests the novel possibility of co-occurrence of complex genomic rearrangements in dystrophinopathy.
Similar content being viewed by others
Introduction
Duchenne and Becker muscular dystrophies (DMD/BMD) are the most commonly inherited muscle diseases that are caused by mutations in the dystrophin gene. This gene consists of 79 exons and is the second largest human gene, spanning more than 2.5 Mb on Xp21.2. It is characterized by a number of large introns and at least eight alternative promoter/first exons scattered among the introns.1, 2 Duplications involving one or more exons of the dystrophin gene are the second most common type of mutation in humans, accounting for approximately 5–10% of dystrophinopathy patients, whereas deletions occupy nearly 60% of mutations3, 4, 5 Though the exact organization of the duplicated exons remains uninvestigated in most cases, it is believed that such duplicated rearrangements consist of a simple tandem head-to-tail duplication within the dystrophin gene. This hypothesis has been adopted to explain DMD or BMD phenotypes based on the reading frame rule that is widely accepted.6
However, one duplication that is not in tandem has been reported,5 suggesting that the above hypothesis is not always correct. In addition, duplicated exons were shown to exist in separate regions of the dystrophin gene.5, 7 Recently, two separate duplicated dystrophin exons were found to consist of two tandem head-to-tail duplications by analysis of dystrophin mRNA.8 These observations suggest that duplication mutations are sometimes complex. So far, all duplications have been reported as intragenic events.
The IL1RAPL1 (interleukin-1 receptor accessory protein-like 1) gene, responsible for X-linked mental retardation, spans 1.36 Mb on Xp22.1, nearly 100 kb distal from the dystrophin gene.9, 10 Both dystrophin and IL1RAPL1 genes were found to reside in an identical common fragile site (FRAXC).10 However, little attention has been paid to IL1RAPL1 gene mutation in dystrophinopathy, even though mental retardation is observed in nearly one-third of DMD patients. The co-occurrence of mutations in these two genes has been reported in a contiguous gene deletion syndrome, in which both dystrophin and IL1RAPL1 genes are affected, the deletion beginning and ending within these genes11, 12, 13 Therefore, it is supposed that an unknown mechanism mutates these two separated genes concurrently. Recently, a mechanism of the fork stalling and template switching (FoSTeS) was proposed as a novel mechanism to mutate two separated genes.14, 15, 16 However, any mutation in the dystrophin gene has not been claimed to be caused by FoSTeS, as far as we know.
Here, we report a previously undescribed duplication of the dystrophin gene that is complicated by the insertion of a duplicated part of the IL1RAPL1 gene into the dystrophin duplication site. This complex rearrangement raises the novel possibility that there is a co-occurrence of two gene mutations even in dystrophinopathy.
Materials and methods
Patients
At the Kobe University Hospital, mutation analysis of the dystrophin gene was conducted in more than 400 DMD/BMD patients. In 39 patients, duplications of one or more exons of the dystrophin gene were disclosed. Among them, 15 patients were further examined for their actual exon junction by dystrophin mRNA analysis as described earlier.8 All except one showed exon structure compatible with their genomic duplication. The proband (KUCG447) was a 10-year-old Japanese boy. At the age of 3 years, his serum creatine kinase level was accidentally found to be markedly elevated (34 070 IU l−1) and he was clinically diagnosed as having DMD. At the age of 6 years, he was referred to Kobe University Hospital to be examined for a mutation in the dystrophin gene. He showed Gowers' sign but mental retardation was not indicated. A muscle biopsy at the age of 8 years disclosed no dystrophin staining, confirming a DMD diagnosis. The Ethical Committee of Kobe University Graduate School of Medicine approved this study and consent for this study was obtained from his parents.
Multiplex ligation-dependent probe amplification analysis
DNA was isolated from lymphocytes derived from the patient, his mother, his maternal grandmother and normal individuals by standard phenol-chloroform extraction methods. Multiplex ligation-dependent probe amplification analysis was performed with the P034 and P035 kits from MRC-Holland (Amsterdam, The Netherlands) by the Mitsubishi Medience Corporation (Tokyo, Japan).17 This technique allowed examination of the full extent of any deletions or duplications of exon(s) in the dystrophin gene.
mRNA analysis
Both dystrophin and IL1RAPL1 mRNAs were examined by reverse transcription-PCR as described earlier.18 Briefly, total RNA was isolated from thin-sliced (6 μm) muscle sections of frozen muscle samples using ISOGEN (Nippon Gene, Toyama, Japan). After synthesizing cDNA with reverse transcriptase (Invitrogen, Carlsbad, CA, USA), target fragments were PCR amplified using conditions essentially as described earlier.19 A fragment encompassing the duplication junction between exons 62 and 56 was amplified using a forward primer corresponding to a segment of exon 61 and a reverse primer complementary to a segment of exon 59 (Table 1).
To examine IL1RAPL1 mRNA, a fragment extending from exons 1 to 9 was amplified using a forward primer corresponding to a segment of exon 1 and a reverse primer complementary to a segment of exon 9 (Table 1). The amplified products were purified and directly sequenced using an automated DNA sequencer (model 310; Applied Biosystems, Foster City, CA, USA).
Genomic quantification of IL1RAPL1 exons
Genomic dosage of exons of the IL1RAPL1 gene was assessed by semiquantitative multiplex PCR as described earlier.20 Five fragments encompassing each exon from 2 to 6 of the IL1RAPL1 gene and one fragment encompassing exon 22 of the dystrophin gene were co-amplified in one PCR reaction using six sets of primers (Table 1). PCR products were separated by capillary electrophoresis (Agilent 2001 Bioanalyzer with DNA 1000 Lab Chips, Agilent Technologies, Palo Alto, CA, USA). The amount of each PCR product from IL1RAPL1 exons was quantified by measuring their peak areas and calculating the ratio of their areas to that of dystrophin exon 22.
Determination of insertion breakpoints
In order to define rearrangement junctions in intron sequences, the ends of the duplications were determined by quantitating PCR amplified products from regions of each intron as described earlier.21 The dystrophin and IL1RAPL1 gene sequences were obtained from the human genome sequence (GenBank NT_011757). Five distributed regions of introns 55 and 62 of the dystrophin gene (120 and 63 kb, respectively) and four regions of introns 2 and 5 of the IL1RAPL1 gene (494 and 269 kb, respectively) were each amplified to yield 150–400-bp-long PCR products (data available on request). The genomic dosage of the amplified intron regions was assessed by semiquantitative multiplex PCR as described earlier.20 The insertion breakpoints were assumed roughly in the region between fragments with single and double amounts of the PCR products. Again, distributed fragments within the breakpoint region assumed from the preceding PCR amplification study were PCR amplified to localize the limit between single and double amounts of the PCR products. This process was repeated to narrow down the breakpoint until the breakpoint junction was presumed to be less than 1000 bp (data available on request). Finally, two junction fragments were obtained: one using a forward primer located in dystrophin intron 62 and a reverse primer in IL1RAPL1 intron 2, and the other using a forward primer from IL1RAPL1 intron 5 and a reverse primer from dystrophin intron 55 (Table 1). The amplified products were then purified and directly sequenced.
Genomic sequence analysis
Four genomic regions corresponding to the junctions in dystrophin introns 55 and 61 and IL1RAPL1 introns 2 and 5, and genomic regions encompassing duplicated exons in both dystrophin and IL1RAPL1 genes were amplified using primers derived from the flanking sequences (Table 1). Amplified products were directly sequenced using an automated DNA sequencer (model 310; Applied Biosystems). Dinucleotide repeat markers from intron 59 (DMD1-2c/DMDI) and intron 62 (DI623) were also analyzed to try to disclose allelic differences as described earlier.22, 23
Examination of the insertion junction sequences
DNA sequences encompassing the insertion junction were analyzed for repetitive elements and low-complexity sequences by use of the RepeatMasker web server (www.repeatmasker.org). Alu sequences were annotated according to Batzer's report.24
Results
In 15 duplications in the dystrophin gene, actual exon junctions were disclosed by sequencing of the reverse transcription-PCR product encompassing the junction region and all but one showed junction sequences compatible with genomic results (data not shown). In the index patient, multiplex ligation-dependent probe amplification analysis of the dystrophin gene showed twice the amount of products for 7 exons consisting of exons 56–62, whereas other exons remained normal (data not shown). On the basis of the conventional understanding of multiplex ligation-dependent probe amplification analysis, exons 56–62 were thought to be involved in a tandem head-to-tail duplication, with the 7 duplicated exons inserted between exons 62 and 63 in the patient's dystrophin mRNA. Applying this assumption to the translational reading frame, a premature stop codon appeared in the sequence of the inserted exon 56. Therefore, the patient's genotype matched with the severe DMD phenotype.
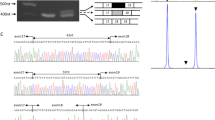
To determine the orientation and location of the duplicated region, the exon structure of dystrophin mRNA was examined. A fragment encompassing the junction between exons 62 and 56 was reverse transcription-PCR amplified using a pair of primers, forward and reverse primers for exon 61 and complementary to exon 59, respectively (Table 1). As a result, the amplification product was obtained from the patient's skeletal muscle RNA, but not from a normal control individual (Figure 1). Unexpectedly, the size of the product (1287 bp) was larger than the expected size of 666 bp. Direct sequencing of the product revealed a novel transcript: sequences of exons 61 and 62 were followed by an unknown 621 bp long sequence, after which sequences of exons 56, 57, 58 and 59 appeared (Figure 1). Both sequences and junctions of dystrophin exons were maintained completely intact. From the order of identified exons in dystrophin mRNA, we concluded that exons 56–62 were tandem head-to-tail duplications, leaving the 621-bp insertion unidentified.
Examination of the duplication junction between dystrophin exons 62 and 56. (a) Schematic description of the examined region of the predicted dystrophin transcript. The boxes represent exons and the numbers inside the box indicate the exon number. Duplicated exons (exons 56–62) are marked by shaded boxes. Horizontal arrows show the locations and directions of the primers that were used for reverse transcription (RT)-PCR amplification. (b) RT-PCR amplified product. RT-PCR product encompassing dystrophin exons 61 to 59 is shown. The product (1287 bp) was obtained from the index patient (right), but the size of the product was larger than the expected 666 bp. However, no amplified product was obtained from the control (left). On the right side, a schematic representation of the exon organization of the amplified fragment is shown. An unknown 621-bp insertion is present between dystrophin exons 62 and 56 (upper); the unknown sequence was found to consist of IL1RAPL1 exons 3 to 5 (lower). (c) Sequence of the junctions between dystrophin exons and the inserted fragment. A part of the sequence of the amplified product is shown. The 3′ terminal sequence of exon 62 (CAA) joined to the 5′ end of the unknown sequence (CCG), and the 3′ end of the unknown sequence (CAG) then joined to the 5′ end of the sequence of exon 56 (GAC). The underlined represents a stop codon (TGA).
A BLAST search of the 621 bp sequence revealed that the sequence was completely identical to a part of the cDNA sequence of the IL1RAPL1 gene (bases 713–1333 of GenBank NM_014271). The inserted 621-bp sequence was found to consist of exons 3, 4 and 5 of the IL1RAPL1 gene, and was inserted in the same direction as the dystrophin exons (Figure 1). These results showed that a novel chimeric dystrophin-IL1RAPL1-dystrophin transcript consisting of dystrophin exons 1–62, IL1RAPL1 exons 3–5 and dystrophin exons 56–79 was produced in the index patient. This transcript encoded a TGA stop codon at the fifth codon in the inserted sequence, consistent with the severe DMD phenotype (Figure 1).
The patient's IL1RAPL1 mRNA was examined by reverse transcription-PCR amplification of a fragment encompassing exons 1–9 of the IL1RAPL1 gene. Surprisingly, this amplification yielded a product with an expected size (1289 bp) and completely normal sequence, including exons 3–5 (Figure 2a). Therefore, we concluded that the IL1RAPL1 gene was intact in the patient's genome.
Analysis of the IL1RAPL1 gene. (a) Reverse transcription (RT)-PCR amplification of the IL1RAPL1 transcript. RT-PCR products encompassing IL1RAPL1 exons 1–9 are shown. One clear product of the same size (1289 bp) was identified in both the index patient and control (left panel). The exon composition of the amplified product is described schematically on the right. Horizontal arrows show the locations and directions of the primers that were used for RT-PCR amplification. (b) Quantification of IL1RAPL1 exons. Capillary electrophoretic patterns of PCR products are shown. Six genomic regions coamplified in one PCR reaction were separated using capillary electrophoresis. Numbers over the peaks indicate exons 2, 3, 4, 5 and 6 of the IL1RAPL1 and exon 22 of the dystrophin gene. The peak area of exons 3, 4 and 5 is nearly doubled in the patient, but normal for exons 2 and 6 (right). The inverted triangle and IS refer to 15- and 1500-bp markers, respectively.
From the above findings, we supposed that an extra copy of exons 3–5 of the IL1RAPL1 gene was present in the patient's genome. We therefore examined the copy number of exons of the IL1RAPL1 gene by semiquantitative PCR. The amount of amplified products from exons 3, 4 and 5 of the IL1RAPL1 gene was nearly twice that of the control, but the same for exons 2 and 6, indicating two copies of exons 3–5 of the IL1RAPL1 gene in the patient's genome (Figure 2b). Taken together with the above results, we concluded that the dystrophin gene acquired an extra copy of the IL1RAPL1 gene between exons 62 and 56 in the same direction as the dystrophin gene.
To confirm the insertion of the IL1RAPL1 gene into the dystrophin gene at the genomic level, insertion breakpoints were examined. At first, the junction between dystrophin intron 62 and IL1RAPL1 intron 2 was narrowed down by repeating semiquantitative PCR amplifications of intron regions (Supplementary Information). Finally, we attempted to PCR amplify the junction fragment between dystrophin intron 62 and IL1RAPL1 intron 2. Although the primer binding sites would be too distant for amplification to occur in the control, the primers should have been in sufficient proximity for amplification to occur in the patient (Figure 3). From the patient, one 995-bp product was obtained. As expected, sequencing of the product revealed the junction sequence between dystrophin intron 62 and IL1RAPL1 intron 2; a multiple sequence alignment revealed that the sequence of intron 62 ended at nt 29066747 (GenBank NT_011757) and joined to nt 26840485 (GenBank NT_011757) of intron 2 of the IL1RAPL1 gene (Figure 3). This showed that dystrophin intron 62 was shortened to 56.7 kb from 62.6 kb, and IL1RAPL1 intron 2 to 242.3 kb from 251.3 kb.
The junction between dystrophin intron 62 and IL1RAPL1 intron 2. Normal sequences of dystrophin intron 62 and IL1RAPL1 intron 2 are aligned in a 5′ to 3′ direction (left to right) at the top and bottom lines, respectively. In the middle, the junction sequence is shown. Vertical lines indicate nucleotide matches. Gaps are indicated by dashes and the inserted nucleotide at the junction is indicated in bold. Vertical arrowheads indicate nucleotide numbers on the X-chromosome (GenBank NT_011757). No significant sequence homology was found around the breakpoint. Exon 1 of G-dystrophin in dystrophin intron 62 is boxed. A GC-box is underlined.
Remarkably, a single C-nucleotide that did not match with either intron sequence was found inserted at this intron junction (Figure 3). Although intron sequences nearby the junction were further examined, neither significant sequence homology between two introns nor specific structures, such as a topoisomerase consensus cleavage site were found. These characteristics indicated that the rearrangement was caused by nonhomologous recombination. Remarkably, it was found that the breakpoint in intron 62 was actually within the first exon of G-dystrophin, a dystrophin isoform transcribed from alternative promoters.25 Thereby, the 194-bp long first exon was disrupted at the 40th nucleotide (Figure 3). In addition, a total of 15 bases (5′-CGGAGCCCGACACGG-3′) (nt 29066825 to 29066810) were found deleted from intron 62 at 65 bp upstream from the junction. Instead, a single T-nucleotide was inserted at this deletion site. The 15-bp deletion removed 10 bp of the 3′ part of one GC-box, constituting a promoter region of G-dystrophin (Figure 3).
Regions covering the junctions in dystrophin intron 62 and IL1RAPL1 intron 2 were amplified. One PCR amplified product specific to each region was obtained from the case as well as from a normal control. Sequencing of the products disclosed completely intact sequences, indicating no genomic changes predisposing the rearrangement. The 15-bp deletion identified upstream of the recombination site in intron 62 was present only in the junction site and not in non-recombined intron. Therefore, the deletion seemed to be an additional genomic rearrangement complicating the duplication mutation. In fact, a G-dystrophin transcript with normal exon content was detected in this patient as in the control, indicating intact intron 62 (data not shown).
As one junction was clearly revealed, we supposed that the junction between IL1RAPL1 intron 5 and dystrophin intron 55 should be present. Using the same strategy as described above (Supplementary Information), the junction fragment was cloned. Finally, the junction fragment was PCR amplified using primers on IL1RAPL1 intron 5 and dystrophin intron 55 and one amplified product was obtained (Figure 4). Sequencing of this product showed a recombination site between the IL1RAPL1 and dystrophin genes; a multiple sequence alignment revealed that the IL1RAPL1 intron 5 sequence ended at nt 27199486 (GenBank NT_011757) and joined to nt 29310447 (GenBank NT_011757) of the dystrophin intron 55 sequence (Figure 4). Between these two sites, 10 nucleotides remained uncategorized in either sequence because of nucleotide mismatches. Therefore, the exact breakpoint of the junction could not be identified. Remarkably, the junction was found to be within an Alu repetitive element in both introns; an Alu Sg element in intron 5 of the IL1RAPL1 gene and an Alu Y element in intron 55 of the dystrophin gene (Figure 4). These two Alu elements were different from each other by 17.7% in their sequences. We decided that this junction was due to a homologous recombination between two Alu repeat sequences. To see any genomic change at junctions in intact introns, regions covering the junction site on IL1RAPL1 intron 5 and dystrophin intron 55 were PCR amplified from the patient's genomic DNA. The sequences of the amplified products were completely normal around the junction in the two introns.
The junction between IL1RAPL1 intron 5 and dystrophin intron 55. Normal sequences of IL1RAPL1 intron 5 and dystrophin intron 55 are shown on the second top and second bottom lines, respectively. In the middle, the sequence across the insertion junction between IL1RAPL1 intron 5 and dystrophin intron 55 is shown. The Alu consensus sequences of Alu Sg and Alu Y are shown on the top and bottom lines, respectively. Vertical lines indicate nucleotide matches. Gaps are indicated by dashes. Before mismatches appear, the IL1RAPL1 intron 5 and dystrophin intron 62 are completely identical to the junction sequences. However, 10 bp, including two mismatches, are not found in either sequence. Vertical arrowheads indicate the rearrangement site with numbers on the X-chromosome (GenBank NT_011757).
These results clearly indicated that a part of the IL1RAPL1 gene was inserted at the junction of the duplication mutation of the dystrophin gene in the same direction as the dystrophin gene. The duplication size of the dystrophin gene was calculated to be 243.7 kb. The size of the inserted fragment of the IL1RAPL1 gene was calculated to be nearly 359.0 kb. Both the patient's mother and maternal grandmother were found to have these two junctions in their genome by PCR amplification of the junction fragment (data not shown), suggesting that the complex duplication had been stably inherited. A total of 30 normal female controls were negative for each junction, suggesting that no part of the rearrangement is polymorphic.
Examination of duplicated exons
As we identified an extra copy of three exons of the IL1RAPL1 gene (Figure 2), we looked for sequence differences between the extra copy and the wild type. The region encompassing the three exons was PCR amplified and directly sequenced. The sequences were completely normal and no heterozygosity was found (data not shown). In addition, all duplicated exons and polymorphic markers in introns 59 and 62 of the dystrophin gene were also analyzed. Their sequences were also completely normal and no heterozygosity was detected (data not shown). These sequencing results strongly suggested that the duplicated fragments were derived from the same genomic source.
In summary, we found the most complex dystrophin duplication mutation yet reported, consisting of a duplication/insertion mutation. The recombination event consisted of two different mechanisms of nonhomologous and Alu-mediated recombination and produced a chimeric transcript.
Discussion
Here, we report a novel complex rearrangement in the dystrophin gene discovered by analyzing the dystrophin transcript. We identified a chimeric dystrophin-IL1RAPL1-dystrophin transcript with IL1RAPL1 exons 3–5 inserted between dystrophin exons 62 and 56. This rearrangement at the genomic level can be roughly summarized as a duplication of exons 56–62 of the dystrophin gene that was accompanied by an insertion of exons 3–5 of the IL1RAPL1 gene. In one study on duplications of the dystrophin gene, one of 118 duplications was reported to be complex, complicating an exon deletion.5 As far as we know, our case is the second example of a complex duplication in the dystrophin gene but the most complicated one. In particular, all duplications have been reported as intragenic events.5, 8, 26 This was the first study to show commitment of other genetic elements to cause duplication in the dystrophin gene. Although the identical duplication of dystrophin exons 56–62 has been reported in two cases (Leiden Muscular Dystrophy pages, www.dmd.nl), analysis of their mRNA has not been carried out. Further study is needed to see whether or not our complicated duplication is an exceptional event.
Although duplications are the second most common mutation in the dystrophin gene, the sequence of the duplication junction has rarely been clarified at the genomic DNA level.5, 27 In this case, a small deletion and several nucleotide changes complicated a large insertion event (Figures 3 and 4). It has been supposed that the sequence or structure of intron 62 may be vulnerable to complex rearrangement.5 Considering that an alternative promoter for G-dystrophin is present within intron 62, this intron should be accessed easily by transcription factors (Figure 3). It is conceivable that unknown structural characteristics promote complex rearrangement in this intron.
It was remarkable that a 359.0-kb fragment of the IL1RAP1 gene located 100 kb apart from the dystrophin gene was found inserted into the duplication junction (Figure 5). An example of transposition of a large fragment from a distant site into the recombination site was reported in the coagulation factor F8 gene located on chromosome Xq28, and the 263-kb duplicated region was shown to have originated 5.2 Mb apart from the inserted site.28 These suggest that the insertion of a large genomic fragment into a far distant region can be a cause of disease.
Schematic description of the genomic rearrangement in the index patient. The dystrophin and IL1RAPL1 genes involved in this rearrangement are described schematically. These two genes are separated by nearly 100 kb on the short arm of the X chromosome (top). The dystrophin gene is transcribed from the centromere (cen) to the telomere (tel), whereas the IL1RAPL1 gene is transcribed in the opposite direction. A 243.7-kb fragment encompassing dystrophin exons 56–62 is duplicated, and at the recombination site another 359.0-kb-long region encompassing exons 3–5 of the IL1RAPL1 gene is inserted (bottom). The rearrangements between dystrophin intron 62 and IL1RAPL1 intron 2 and between IL1RAPL1 intron 5 and dystrophin intron 55 are through nonhomologous and Alu-mediated recombination, respectively. The rearrangement is illustrated by its predicted order and origins (middle). The bold horizontal arrows indicate the dystrophin and IL1RAPL1 genes and their transcriptional direction. The duplicated region is boxed with dotted lines. Open, shaded and dotted boxes indicate dystrophin exons 56–62, IL1RAPL1 exons 3–5 and the alternative G-dystrophin promoter/first exon (G1), respectively. Numbers over bars indicate the exon number of the respective genes. Ovals represent Alu repeat sequences (Alu). Parentheses indicate the size of the regions. Size is not to scale. Vertical arrows indicate junctions.
Considering that both dystrophin and IL1RAPL1 genes are present in the same common fragile site (FRAXC),10 it is reasonable to consider co-occurrence of mutations in these two genes11, 12, 13 Production of a chimeric dystrophin-IL1RAPL1 transcript, however, has been shown in one DMD patient who had a deletion encompassing ∼1.6 Mb from the IL1RAPL1 gene to the dystrophin gene.12 Our case is the second to show the production of a chimeric dystrophin-IL1RAPL1 transcript (Figure 1). Remarkably, both rearrangements producing a chimeric dystrophin-IL1RAPL1 transcript occurred in the Alus of IL1RAPL1 intron 5 located only 50 kb apart.
Recombination mechanisms, such as nonallelic homologous recombination and nonhomologous end joining, have been shown to underlie rearrangements causing genomic disorders,16, 29 with the former accounting for the majority. It has been suggested that duplications within the dystrophin gene are primarily caused by unequal sister-chromatid exchange.30, 31 However, synthesis-dependent nonhomologous end joining has been proposed as a cause of duplications, resulting in a tandem duplication at the site of a double-strand break.5 Large tandem duplications are well-known causes of human genetic disease.32 Especially in Pelizaeus–Merzbacher disease, an X-linked dysmyelinating disorder, nonrecurrent duplication of the dosage-sensitive PLP1 gene is known as the most common cause.33 Recently, it was proposed that complex duplications of the proteolipid protein 1 (PLP1) gene are due to the replication-based mechanism of ‘replication folk stalling and template switching (FoSTeS),’ as these duplications could be explained by neither nonallelic homologous recombination nor a simple nonhomologous end joining recombination mechanism.14 In our case, it is better to consider this possibility as the underlying mechanism for the complex genomic rearrangement (Figure 5).
As mutations of the IL1RAPL1 gene have been identified in X-linked mental retardation,34, 35, 36 our finding that three exons of the IL1RAPL1 gene were transposed into the dystrophin gene suggested the possibility of X-linked mental retardation caused by their deletion. Accordingly, there is one case of a three-exon deletion described in the literature resulting in mental retardation.34 Therefore, we strongly suspect that concomitant mutations in the IL1RAPL1 gene and dystrophin gene account for both DMD and mental retardation. Further studies will elucidate the co-occurrence of mutations in both these genes and help explain why mental retardation is a complicating factor in DMD patients.
References
Ahn, A. H. & Kunkel, L. M. The structural and functional diversity of dystrophin. Nat. Genet. 3, 283–291 (1993).
Nishio, H., Takeshima, Y., Narita, N., Yanagawa, H., Suzuki, Y., Ishikawa, Y. et al. Identification of a novel first exon in the human dystrophin gene and of a new promoter located more than 500 kb upstream of the nearest known promoter. J. Clin. Invest. 94, 1037–1042 (1994).
Hu, X. Y., Ray, P. N., Murphy, E. G., Thompson, M. W. & Worton, R. G. Duplicational mutation at the Duchenne muscular dystrophy locus: its frequency, distribution, origin, and phenotypegenotype correlation. Am. J. Hum. Genet. 46, 682–695 (1990).
White, S., Kalf, M., Liu, Q., Villerius, M., Engelsma, D., Kriek, M. et al. Comprehensive detection of genomic duplications and deletions in the DMD gene, by use of multiplex amplifiable probe hybridization. Am. J. Hum. Genet. 71, 365–374 (2002).
White, S. J., Aartsma-Rus, A., Flanigan, K. M., Weiss, R. B., Kneppers, A. L., Lalic, T. et al. Duplications in the DMD gene. Hum. Mutat. 27, 938–945 (2006).
Monaco, A. P., Bertelson, C. J., Liechti-Gallati, S., Moser, H., Kunkel, L. M. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics 2, 90–95 (1988).
Janssen, B., Hartmann, C., Scholz, V., Jauch, A. & Zschocke, J. MLPA analysis for the detection of deletions, duplications and complex rearrangements in the dystrophin gene: potential and pitfalls. Neurogenetics 6, 29–35 (2005).
Zhang, Z., Takeshima, Y., Awano, H., Nishiyama, A., Okizuka, Y., Yagi, M. et al. Tandem duplications of two separate fragments of the dystrophin gene in a patient with Duchenne muscular dystrophy. J. Hum. Genet. 53, 215–219 (2008).
Jin, H., Gardner, R. J., Viswesvaraiah, R., Muntoni, F. & Roberts, R. G. Two novel members of the interleukin-1 receptor gene family, one deleted in Xp22.1-Xp21.3 mental retardation. Eur. J. Hum. Genet. 8, 87–94 (2000).
McAvoy, S., Ganapathiraju, S., Perez, D., James, C. & Smith, D. DMD and IL1RAPL1: two large adjacent genes localized within a common fragile site (FRAXC) have reduced expression in cultured brain tumors. Cytogenet. Genome Res. 119, 196–203 (2007).
Niakan, K., Zhang, Y., Huang, B. & McCabe, E. Complex glycerol kinase deficiency;defining the deletion breakpoints in the original patient in this contiguous gene syndrome. Am. J. Hum. Genet. 69, 632 (2001).
Wheway, J. M., Yau, S. C., Nihalani, V., Ellis, D., Irving, M., Splitt, M. et al. A complex deletion-inversion-deletion event results in a chimeric IL1RAPL1-dystrophin transcript and a contiguous gene deletion syndrome. J. Med. Genet. 40, 127–131 (2003).
Klein, R. D., Thorland, E. C., Gonzales, P. R., Beck, P. A., Dykas, D. J., McGrath, J. M. et al. A multiplex assay for the detection and mapping of complex glycerol kinase deficiency. Clin. Chem. 52, 1864–1870 (2006).
Lee, J., Carvalho, C. & Lupski, J. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131, 1235–1247 (2007).
Branzei, D. & Foiani, M. Template switching: from replication fork repair to genome rearrangements. Cell 131, 1228–1230 (2007).
Gu, W., Zhang, F. & Lupski, J. R. Mechanisms for human genomic rearrangements. Pathogenetics 1, 4 (2008).
Lalic, T., Vossen, R., Coffa, J., Schouten, J., Guc-Scekic, M., Radivojevic, D. et al. Deletion and duplication screening in the DMD gene using MLPA. Eur. J. Hum. Genet. 13, 1231–1234 (2005).
Matsuo, M., Masumura, T., Nishio, H., Nakajima, T., Kitoh, Y., Takumi, T. et al. Exon skipping during splicing of dystrophin mRNA precursor due to an intraexon deletion in the dystrophin gene of Duchenne muscular dystrophy Kobe. J. Clin. Invest. 87, 2127–2131 (1991).
Tran, V. K., Takeshima, Y., Zhang, Z., Habara, Y., Haginoya, K., Nishiyama, A. et al. A nonsense mutation-created intraexonic splice site is active in the lymphocytes, but not in the skeletal muscle of a DMD patient. Hum. Genet. 120, 737–742 (2007).
Tran, V. K., Zhang, Z., Yagi, M., Nishiyama, A., Habara, Y., Takeshima, Y. et al. A novel cryptic exon identified in the 3′ region of intron 2 of the human dystrophin gene. J. Hum. Genet. 50, 425–433 (2005).
Silao, C. L., Padilla, C. D. & Matsuo, M. A novel deletion creating a new terminal exon of the dihydrolipoyl transacylase gene is a founder mutation of Filipino maple syrup urine disease. Mol. Genet. Metab. 81, 100–104 (2004).
Powell, J. F., Fodor, F. H., Cockburn, D. J., Monaco, A. P. & Craig, I. W. A dinucleotide repeat polymorphism at the DMD locus. Nucleic. Acids Res. 19, 1159 (1991).
Matsumoto, T. & Niikawa, N. Eight novel microsatellite markers in the 3′ region of the dystrophin gene useful for diagnosis of Duchenne muscular dystrophy. Prenat. Diagn. 24, 1014–1015 (2004).
Batzer, M. A., Deininger, P. L., Hellmann-Blumberg, U., Jurka, J., Labuda, D., Rubin, C. M. et al. Standardized nomenclature for Alu repeats. J. Mol. Evol. 42, 3–6 (1996).
Hugnot, J. P., Gilgenkrantz, H., Vincent, N., Chafey, P., Morris, G. E., Monaco, A. P. et al. Distal transcript of the dystrophin gene initiated from an alternative first exon and encoding a 75-kDa protein widely distributed in nonmuscle tissues. Proc. Natl Acad. Sci. USA 89, 7506–7510 (1992).
Gualandi, F., Rimessi, P., Trabanelli, C., Spitali, P., Neri, I., Patarnello, T. et al. Intronic breakpoint definition and transcription analysis in DMD/ BMD patients with deletion/ duplication at the 5′ mutation hot spot of the dystrophin gene. Gene 370, 26–33 (2006).
Weiss, C., Jakubiczka, S., Huebner, A., Klopocki, E., Kress, W., Voit, T. et al. Tandem duplication of DMD exon 18 associated with epilepsy, macroglossia, and endocrinologic abnormalities. Muscle Nerve 35, 396–401 (2007).
Sheen, C., Jewell, U., Morris, C., Brennan, S., Férec, C., George, P. et al. Double complex mutations involving F8 and FUNDC2 caused by distinct break-induced replication. Hum. Mutat. 28, 1198–1206 (2007).
Shaw, C. & Lupski, J. Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Hum. Mol. Genet. R 1, R57–R64 (2004).
Hu, X. Y., Burghes, A. H., Bulman, D. E., Ray, P. N. & Worton, R. G. Evidence for mutation by unequal sister chromatid exchange in the Duchenne muscular dystrophy gene. Am. J. Hum. Genet. 44, 855–863 (1989).
Hu, X., Ray, P. & Worton, R. Mechanisms of tandem duplication in the Duchenne muscular dystrophy gene include both homologous and nonhomologous intrachromosomal recombination. EMBO J. 10, 2471–2477 (1991).
Chen, J. M., Chuzhanova, N., Stenson, P. D., Ferec, C. & Cooper, D. N. Complex gene rearrangements caused by serial replication slippage. Hum. Mutat. 26, 125–134 (2005).
Woodward, K. J., Cundall, M., Sperle, K., Sistermens, E. A., Ross, M., Howell, G. et al. Heterogeneous duplications in patients with Pelizaeus-Merzbacher disease suggest a mechanism of coupled homologous and nonhomologous recombination. Am. J. Hum. Genet. 77, 966–987 (2005).
Carrie, A., Jun, L., Bienvenu, T., Vinet, M. C., McDonell, N., Couvert, P. et al. A new member of the IL-1 receptor family highly expressed in hippocampus and involved in X-linked mental retardation. Nat. Genet. 23, 25–31 (1999).
Sasaki, R., Inamo, Y., Saitoh, K., Hasegawa, T., Kinoshita, E. & Ogata, T. Mental retardation in a boy with congenital adrenal hypoplasia: a clue to contiguous gene syndrome involving DAX1 and IL1RAPL. Endocr. J. 50, 303–307 (2003).
Zhang, Y. H., Huang, B. L., Niakan, K. K., McCabe, L. L., McCabe, E. R. & Dipple, K. M. IL1RAPL1 is associated with mental retardation in patients with complex glycerol kinase deficiency who have deletions extending telomeric of DAX1. Hum. Mutat. 24, 273 (2004).
Acknowledgements
We thank Prof James R Lupski for his kind and critical comments in the preparation of this article. We thank Ms Misato for secretarial assistance. This work was supported by a Grant-in-Aid for Scientific Research (B) and a Grant-in-Aid for Exploratory Research from the Japan Society for the Promotion of Science, by a Health and Labour Sciences Research Grant for Research on Psychiatric and Neurological Diseases and Mental Health, and a Research Grant for Nervous and Mental Disorders, from the Ministry of Health, Labour and Welfare of Japan and by a grant from the New Energy and Industrial Technology Development Organization.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website (http://www.nature.com/jhg)
Supplementary information
Rights and permissions
About this article
Cite this article
Zhang, Z., Yagi, M., Okizuka, Y. et al. Insertion of the IL1RAPL1 gene into the duplication junction of the dystrophin gene. J Hum Genet 54, 466–473 (2009). https://doi.org/10.1038/jhg.2009.63
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2009.63