Abstract
Isolated cytochrome c oxidase (COX) deficiency (MIM#220110) is a relatively common biochemical finding in pediatric patients with mitochondrial disorder. It has been associated with different clinical phenotypes ranging from isolated myopathy to severe multisystem disorder. It is a genetically heterogeneous trait, and the most frequent genetic defects affect SURF1 and SCO2, two genes required for COX assembly. However, a significant proportion of patients lacks mutation in these genes and in other known genes that require COX biogenesis. COX18 is a novel COX assembly gene required for membrane insertion of the C-terminal portion of COX subunit II. We have studied 29 pediatric patients with isolated COX deficiency in the skeletal muscle associated with different clinical phenotypes. Mutations in SURF1, SCO2, SCO1, COX10, COX15 and in mitochondrial DNA, had been ruled out earlier. The COX18 gene was analyzed using a PCR-single-stranded conformation polymorphism (PCR-SSCP) protocol, and in 15 patients, the analysis was repeated by direct sequencing. No pathogenic mutations were detected in our cohort of patients indicating that COX18 mutations may be very rare or associated with other phenotypes than isolated COX deficiency in infancy.
Similar content being viewed by others
Introduction
Cytochrome c oxidase (COX), the terminal component of the mitochondrial electron transport chain, comprises 13 protein subunits, 3 encoded by mitochondrial DNA (mtDNA) and 10 by nuclear DNA (nDNA).1 COX requires other ancillary proteins for its biogenesis, encoded by COX assembly genes, which synthesize the prosthetic groups, transport and insert the metal cofactors, as well as assemble the different subunits to form a holoenzyme.2 In humans, isolated COX deficiency (MIM#220110) is a common cause of mitochondrial dysfunction in infancy and is clinically and genetically heterogeneous.3 Mutations in the mtDNA-encoded subunits are rare; most defects are inherited as autosomal recessive traits, but mutations in nDNA-encoded subunits have been found only in one patient.4 However, mutations have been detected in six COX assembly genes, namely SURF1, SCO1, SCO2, COX10, COX15 and LRPPRC (see Barrientos et al.2 for individual references). SURF1 and SCO2 account for the majority of reported defects;5 mutations in COX10, COX15 and SCO1 are less frequent, whereas those in LRPPRC are specific to Leigh Syndrome French Canadian type, a distinct clinical entity found (almost) exclusively in patients from the Saguenay-Lac Saint-Jean region of Canada.6 Nevertheless, it is not possible to find any defect in a large proportion of patients.7, 8
We have identified a novel human COX assembly gene, COX18,9 which encodes a mitochondrial protein of 333 amino acids. It belongs to the Alb3–Oxa1–YidC protein family,10 which is required for the processing of transmembrane proteins in mitochondria, chloroplasts and bacteria. In yeast, Cox18p is required for the insertion of the C-terminal tail of COX subunit II into the mitochondrial inner membrane. COX18null Saccharomyces cerevisiae show severe isolated COX deficiency, and the human gene can complement the defect.11, 12 Therefore, we tested whether mutations of human COX18 could play a role in patients with isolated COX deficiency.
Materials and methods
Patients
We have studied 29 patients, of different ethnical background, who had biochemically documented isolated COX deficiency in muscle and different clinical phenotypes. A total of 21 patients had been screened earlier for mutations in the 3 mitochondrial COX subunits, the 22 mtDNA transfer RNA (tRNA) genes and in 7 COX assembly genes, namely SCO1, SCO2, SURF1, COX10, COX11, COX15 and COX17 (patients 3–7, 9–11, 13–16, 19–22, 24–27 and 29–30 as reported by Sacconi et al.7). Eight previously unpublished patients were included in the study. Five presented with encephalomyopathy and liver failure but lacked radiological features of the LS. Two patients had LS, and two had encephalomyopathy without LS. Muscle biopsies had been obtained with the informed consent of the patients' legal tutors, and were analyzed as reported.7 All had isolated COX deficiency in muscle, and histochemistry showed a diffuse reduction in COX staining. They tested negative for mutations in COX10, SCO1, SCO2, SURF1, COX15, mitochondrial tRNA genes, mitochondrial COX subunits and in COX6B1. Eight of the nine patients with hepatic involvement had also been screened for mutations in LPPRC. Phenotypes have been summarized in Table 1.
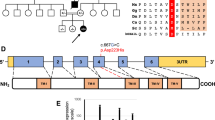
The six coding exons of COX18 and intron–exon boundaries amplified using primers and conditions are summarized in Table 2. The amplicons were analyzed by single-stranded conformation polymorphism (SSCP) using a reported protocol.13 Owing to its size, exon 1 was analyzed by direct sequencing in all patients. Fragments showing abnormal patterns were sequenced as described.13
In a subset of patients, the whole coding region of COX18 was also sequenced directly using the same primer set.
Results and discussion
Single-stranded conformation polymorphism analysis did not detect pathogenic mutations in our cohort of patients. As COX18 is highly expressed in the liver, we focused on the group of patients with predominant hepatic involvement. To exclude that, our protocol had overlooked some mutations (we estimate its sensitivity to be ∼80–90%); we sequenced the whole coding region of COX18 in these nine patients, and also in four patients with LS, in one patient with encephalomyopathy and in the patient with isolated myopathy. Again, no pathogenic mutations were detected. It should be noted that there were no discrepancies between the results of the SSCP analysis and of direct sequencing in these patients.
We considered that COX18 could be an interesting candidate gene in patients with isolated COX deficiency, because COX18null S. cerevisiae strains retain some residual COX activity,11 and this fact may be important for avoiding the embryonic lethality-associated complete mitochondrial respiration defects.14
Our analysis did not detect pathogenic mutations in this gene, even in the 15 patients who were analyzed by direct sequencing. COX18 mutations might be very rare or associated with other phenotypes than those that we studied.
Mutation screening in these patients is problematic because of the large (and still growing) number of candidate genes. The clinical picture is often not helpful in directing the genetic analysis, because there is a significant overlap between phenotypes associated with specific gene defects (for example, hypertrophic cardiomyopathy is a feature of SCO2, COX10 and COX15 mutations2), and because of the phenotypic variability of mutations in individual genes, such as COX1015 and COX15.16 Moreover, SURF1 defects, which are usually associated with LS, may also present with different clinical phenotypes.17, 18
Most patients with COX deficiency are isolated cases and therefore, it is impossible to use traditional genetic techniques, such as linkage or homozygosity mapping, to direct molecular analyses. Microcell-mediated chromosome transfer has been used successfully for identifying specific gene defects,19 but it is a complex and manpower-intensive procedure and is not suitable for analyzing large cohorts of patients. Simpler methods have been developed on the basis of functional complementation of cultured fibroblasts using specific lentiviral vectors;15 however, for most of our patients, the muscle biopsy fragment was the only biological material available for our study. Therefore, we stress the importance of collecting skin fibroblasts in all patients in whom a respiratory chain defect is suspected.
Novel high-throughput techniques will soon become available, and it will be possible to rapidly analyze a large panel of genes in individual patients at reasonable costs. Therefore, it is important to identify other human COX assembly genes as only a portion of the at least 30 different yeast genes involved in COX biogenesis20 have been characterized in humans.
References
Michel, H. The mechanism of proton pumping by cytochrome c oxidase. Proc. Natl. Acad. Sci. USA 95, 12819–12824 (1998).
Barrientos, A., Gouget, K., Horn, D., Soto, I. C., Fontanesi, F. Suppression mechanisms of COX assembly defects in yeast and human: insights into the COX assembly process. Biochim. Biophys. Acta. 1793, 97–107 (2009).
Shoubridge, E. A. Cytochrome c oxidase deficiency. Am. J. Med. Genet. 106, 46–52 (2001).
Massa, V., Fernandez-Vizarra, E., Alshahwan, S., Bakhsh, E., Goffrini, P., Ferrero, I. et al. Severe infantile encephalomyopathy caused by a mutation in COX6B1, a nucleus-encoded subunit of cytochrome c oxidase. Am. J. Hum. Genet. 82, 1281–1289 (2008).
Bohm, M., Pronicka, E., Karczmarewicz, E., Pronicki, M., Piekutowska-Abramczuk, D., Sykut-Cegielska, J. et al. Retrospective, multicentric study of 180 children with cytochrome c oxidase deficiency. Pediatr. Res. 59, 21–26 (2006).
Mootha, V. K., Lepage, P., Miller, K., Bunkenborg, J., Reich, M., Hjerrild, M. et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc. Natl Acad. Sci. USA 100, 605–610 (2003).
Sacconi, S., Salviati, L., Sue, C. M., Shanske, S., Davidson, M. M., Bonilla, E. et al. Mutation screening in patients with isolated cytochrome c oxidase deficiency. Pediatr. Res. 53, 224–230 (2003).
Coenen, M. J., Smeitink, J. A., Pots, J. M., van Kaauwen, E., Trijbels, F. J., Hol, F. A. et al. Sequence analysis of the structural nuclear encoded subunits and assembly genes of cytochrome c oxidase in a cohort of 10 isolated complex IV-deficient patients revealed five mutations. J. Child Neurol. 21, 508–511 (2006).
Sacconi, S., Trevisson, E., Pistollato, F., Baldoin, M. C., Rezzonico, R., Bourget, I. et al. hCOX18 and hCOX19: two human genes involved in cytochrome c oxidase assembly. Biochem. Biophys. Res. Commun. 337, 832–839 (2005).
Kuhn, A., Stuart, R., Henry, R. & Dalbey, R. E. The Alb3/Oxa1/YidC protein family: membrane-localized chaperones facilitating membrane protein insertion? Trends Cell Biol. 13, 510–516 (2003).
Souza, R. L., Green-Willms, N. S., Fox, T. D., Tzagoloff, A. & Nobrega, F. G. Cloning and characterization of COX18, a Saccharomyces cerevisiae PET gene required for the assembly of cytochrome oxidase. J. Biol. Chem. 275, 14898–14902 (2000).
Gaisne, M. & Bonnefoy, N. The COX18 gene, involved in mitochondrial biogenesis, is functionally conserved and tightly regulated in humans and fission yeast. FEMS Yeast Res. 6, 869–882 (2006).
Salviati, L., Sacconi, S., Mancuso, M., Otaegui, D., Camano, P., Marina, A. et al. Mitochondrial DNA depletion and dGK gene mutations. Ann. Neurol. 52, 311–317 (2002).
Vempati, U. D., Diaz, F., Barrientos, A., Narisawa, S., Mian, A. M., Millan, J. L. et al. Role of cytochrome c in apoptosis: increased sensitivity to tumor necrosis factor alpha is associated with respiratory defects but not with lack of cytochrome c release. Mol. Cell Biol. 27, 1771–1783 (2007).
Antonicka, H., Leary, S. C., Guercin, G. H., Agar, J. N., Horvath, R., Kennaway, N. G. et al. Mutations in COX10 result in a defect in mitochondrial heme A biosynthesis and account for multiple, early-onset clinical phenotypes associated with isolated COX deficiency. Hum. Mol. Genet. 12, 2693–2702 (2003).
Bugiani, M., Tiranti, V., Farina, L., Uziel, G. & Zeviani, M. Novel mutations in COX15 in a long surviving Leigh syndrome patient with cytochrome c oxidase deficiency. J. Med. Genet. 42, e28 (2005).
Salviati, L., Freehauf, C., Sacconi, S., DiMauro, S., Thoma, J. & Tsai, A. C. Novel SURF1 mutation in a child with subacute encephalopathy and without the radiological features of Leigh Syndrome. Am. J. Med. Genet. A 128A, 195–198 (2004).
Tay, S. K., Sacconi, S., Akman, H. O., Morales, J. F., Morales, A., De Vivo, D. C. et al. Unusual clinical presentations in four cases of Leigh disease, cytochrome c oxidase deficiency, and SURF1 gene mutations. J. Child Neurol. 20, 670–674 (2005).
Zhu, Z., Yao, J., Johns, T., Fu, K., De Bie, I., Macmillan, C. et al. SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat. Genet. 20, 337–343 (1998).
Tzagoloff, A. & Dieckmann, C. L. PET genes of Saccharomyces cerevisiae. Microbiol. Rev. 54, 211–225 (1990).
Acknowledgements
This work has been supported by Telethon Italy Grant GGP06256 and by a grant from Association Francaise Contre les Myopathies. We are grateful to Dr Salvatore DiMauro, Columbia University, Department of Neurology, New York, for providing the patients samples for this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sacconi, S., Salviati, L. & Trevisson, E. Mutation analysis of COX18 in 29 patients with isolated cytochrome c oxidase deficiency. J Hum Genet 54, 419–421 (2009). https://doi.org/10.1038/jhg.2009.36
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2009.36
Keywords
This article is cited by
-
A biallelic variant in COX18 cause isolated Complex IV deficiency associated with neonatal encephalo-cardio-myopathy and axonal sensory neuropathy
European Journal of Human Genetics (2023)