Abstract
An actinobacterial strain, DCWR9-8-2T, was isolated from a leaf of Thai upland rice (Oryza sativa) collected in Chumporn province, Thailand. Strain DCWR9-8-2T is Gram-stain-positive aerobic bacteria that produce single spores directly on the vegetative hypha. Cell wall peptidoglycan of this strain exhibits meso-diaminopimelic acid and glycine, the reducing sugars of whole-cell hydrolysate are arabinose, glucose, ribose, xylose and small amount of mannose. The phospholipid profiles in the membrane are comprised of phosphatidylethanolamine, diphosphatidylglycerol, phosphatidylglycerol, phosphatidylinositol, phosphatidylinositol mannosides. The major menaquinones are MK-9(H4) and MK-10(H6). The diagnostic cellular fatty acids are iso-C16:0 and iso-C15:0. The G+C content of the genomic DNA is 72.5 mol%. The result of 16S rRNA sequence analysis of the strain revealed that this strain was closely related to Micromonospora auratinigra TT1-11T (99.25%). On the other hand, the result of gyrB gene sequence analysis revealed that this strain was closed to M. eburnea JCM 12345T (96.30%). In addition, a combination of DNA–DNA hybridization results and some phenotypic properties supported that this strain should be judged as a novel species of the genus Micromonospora, for which the name M. endophytica sp. nov. is proposed. The type strain is DCWR9-8-2T (=BCC 67267T=NBRC 110008T).
Similar content being viewed by others
Introduction
Orskov1 is the first scientist that proposed the actinobacteria, namely Micromonospora. All members of this genus produced single non-motile spores directly on the substrate mycelium. This organism cannot produce aerial mycelium during its life cycle. Cell wall peptidoglycan of this organism contains glutamic acid, glycine, alanine and diaminopimelic acids. Peptidoglycan type (Schleifer and Kandler2) of Micromonospora is A1γ. The acyl type of cell wall muramic acid is glycolyl. Diagnostic reducing sugars of cell hydrolysates are xylose and arabinose. Characteristic phospholipids are phosphatidylethanolamine. The major cellular fatty acids are iso-C16:0, iso-C15:0, iso-C17:0, anteiso-C15:0, anteiso-C17:0 and 10-methyl-C17:0. Mycolic acids are absent. The range of DNA G+C contents (mol%) is 71–73. Micromonospora is well known as antibiotic producers. Several antibiotics used in the present time, that is, gentamicin,3 lomaiviticins,4 lupinacidin,5 maklamicin,6 are produced by the members of this genus. Nowadays, 61 species have been characterized as members of the genus Micromonospora. Here, we described the taxonomic characterization of a novel endophytic Micromonospora strain DCWR9-8-2T, which was isolated from a leaf of Thai upland rice.
Materials and Methods
Strain DCWR9-8-2T was isolated from a leaf of upland rice collected from Chumporn province, Thailand. A Leaf sample was rinsed with tap water for three times and was surface-sterilized with 70% ethyl alcohol for 5 min and subsequently with absolute ethyl alcohol for 1 min. A treated leaf was dried on sterilized filter paper and then was treated with 6% sodium hypochlorite in distilled water for 2 min. After that, the leaf tissue was rinsed with sterilized distilled water for five times and the surface-sterilized leaf was ground in sterilized distilled water. Two hundred microlitres of the plant solution was spread on starch casein agar (0.1% sodium caseinate, 1% soluble starch, 0.03% K2HPO4, 1.8% agar, pH 7.0–7.5) supplemented with 25 mg l−1 nalidixic acid and 100 mg l−1 nystatin and incubated at 30 °C for 21 days. The final rinsing water was spread on starch casein agar as the control plates. The colony of strain DCWR9-8-2T was selected and purified on ISP 2 medium (International Streptomyces Project, ISP 2 medium).7 Strain DCWR9-8-2T was grown on starch casein agar medium at 30 °C for 21 days. Then, the colonies were observed by scanning electron microscopy (model LEO/1455VP). The scanning electron microscopic samples of strain DCWR9-8-2T were prepared as described previously.8
Several standard methods were used for the phenotypic determination. Cultural properties were examined by using 14-day cultures grown at 30 °C on various agar media. The determining color designations was judged using the ISCC–NBS Color Charts standard sample no. 2106.9 The determination of the carbon utilization of the strain was done using International Streptomyces Project medium no.9 (ISP9 medium) supplemented with a final concentration of 1% of the carbon sources. The basal medium recommended by Gordon et al.10 were used for the examination of the decomposition of various compounds and acid production from carbon sources. The test for growth at various temperatures (10–50 °C), different sodium chloride concentration (0–12% NaCl) and pH 4–12 was examined on ISP 2 medium. The desired pH was adjusted using sterile solutions of citric acid/Na2HPO4 (for pH 4.0–5.0), Na2HPO4/NaH2PO4 buffer (for pH 6.0–8.0), NaHCO3/Na2CO3 buffer (for pH 9.0–10.0), Na2HPO4/ NaOH buffer (pH 11.0) and KCl/NaOH buffer (for pH 12.0–13.0). The culture media and methods described by Arai11 and Williams and Cross12 were used for determination of starch hydrolysis, peptonization of milk, gelatin liquefaction and the reduction of nitrate.
Cells of strain DCWR9-8-2T were grown in ISP 2 broth on rotary shaker (200 r.p.m.) at 30 °C for 5 days and used for chemotaxonomic analyses. Cell wall peptidoglycan was prepared by the methods of Kawamoto et al.,13 and the isomer of diaminopimelic acid was analyzed by the method of Staneck and Roberts.14 The acyl group in the peptidoglycan was performed as described by Uchida and Aida.15 The reducing sugars of cell hydrolysate were determined by the method of Komagata and Suzuki.16 The phospholipid of the cell membranes were analyzed by the method of Minnikin et al.17 The analysis of cellular fatty acid profile was performed by gas liquid chromatography according to the instructions of the Microbial Identification System (MIDI, version 6.0, Newark, DE, USA) (Sasser;18 Kämpfer and Kroppenstedt19) with the ACTIN1 MIDI database. Menaquinones were extracted by the method of Collins et al.20 and were examined by HPLC equipped with a Cosmosil 5C18 column (4.6 × 150 mm; Nacalai Tesque, Kyoto, Japan). The elution solvent was a mixture of methanol and 2-propanol (2:1, v/v). The presence of mycolic acids was investigated using the method of Minnikin et al.21
Genomic DNA of strain DCWR9-8-2T used for polymerase chain reaction, the G+C content analysis and DNA–DNA hybridization was prepared according to the method of Tamaoka.22 The G+C content (mol %) was analyzed using the HPLC method of Tamaoka and Komagata.23 Lambda DNA (Invitrogen, USA) was used as the standard. DNA–DNA hybridization was determined as described by Ezaki et al.24 DNA–DNA relatedness (%) was analyzed by the colorimetric method of Verlander.25 The 16S rRNA gene fragment was amplified as described by Suriyachadkun et al.26
Amplification and sequencing of the gyrB gene was performed according to the previously described method.27 The sequence of the 16S rRNA gene was obtained by using the universal primers.28 The pairwise alignment and the values for sequence similarity of the 16S rRNA gene and the gyrB gene sequence of strain DCWR9-8-2T were performed using the EzTaxon server29 and CLUSTAL X program (Thompson et al.),30 respectively. Partial sequences of the 16S rRNA gene and the gyrB gene of recognized Micromonospora species were obtained from the GenBank/EMBL/DDBJ databases for multiple alignment analyses using the CLUSTAL W programme, version 1.81.31 Prior to the construction of a phylogenetic tree, the alignment of 16S rRNA gene and the gyrB gene sequences was manually verified and adjusted. The phylogenetic analysis was constructed using the neighbor-joining32 and maximum-likelihood33 methods in the MEGA 5 software.34 The evolutionary distances of the neighbor-joining tree were calculated by using the Kimura two-parameter model.35 The confidence values of branches in the tree were determined using bootstrap analyses36 with 1000 repeats.
Results and Discussion
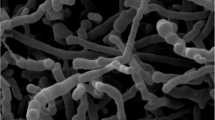
Strain DCWR9-8-2T exhibited a range of phenotypic and chemotaxonomic properties that was consistent with their classification in the genus Micromonospora.37 The strain grew well on ISP 2, ISP 3 and ISP 6, moderately on nutrient agar, weakly on ISP 4, ISP 5, ISP 7, czapek’s sucrose agar and glucose-asparagine agar (Table 1). Strain DCWR9-8-2T formed branched substrate mycelia but aerial mycelia were not produced in all media test. Spores of this strain were borne singly on the substrate mycelia and the spore surface was smooth (Figure 1). The color of substrate mycelia on these media was pale orange yellow to deep orange yellow. The orange yellow soluble pigment was produced on ISP2 and ISP 3. The range of pH and temperature for growth of strain DCWR9-8-2T were pH 5–10 and 20–45 °C, respectively. The maximum NaCl tolerance was 2% (w/v). Other phenotypic characteristics are given in Table 2 and in the species description. Chemotaxonomic properties of strain DCWR9-8-2T were similar to those of members of the genus Micromonospora. This organism contained meso-diaminopimelic acid as the diagnostic diamino acid in the cell wall peptidoglycan; the acyl type of cell wall muramic acid was glycolyl. Arabinose, glucose, ribose, xylose and small amount of mannose were observed as reducing sugars in cell hydrolysates. The menaquinones in cell of strain DCWR9-8-2T were MK-9(H4) (27.0%) and MK-10(H6) (25.9%); substantial amounts of MK-10(H4) (15.3%), MK-9(H2) (13.1%), MK-10(H8) (9.6%), MK-10(H2) (8.8%) and MK-9(H6) (0.2%) were also present. The polar lipids in cell membrane were diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, phosphatidylinositol, phosphatidylinositol mannosides, five unidentified phospholipids and two unidentified lipids (Supplementary Figure S2). Mycolic acids were not observed. The predominant cellular fatty acids (⩾5%) were iso-C16:0, iso-C15:0, anteiso-C17:0, anteiso-C15:0 and iso-C17:0 (Supplementary Table S1). The DNA base compositions of strain was 72.5 mol%.
The result of pairwise alignment analysis of the partial 16S rRNA gene sequence of strain DCWR9-8-2T (1502 nt) obtained from the EzTaxon server29 showed that strain DCWR9-8-2T is a member of the genus Micromonospora. The highest 16S rRNA gene sequence similarity value was observed with M. auratinigra TT1-11T (99.25%) followed by M. echinospora ATCC 15837T (99.15%), M. chaiyaphumensis MS5-1T (98.96%), M. pattaloongensis TJ2-2T (98.91%) and M. eburnea LK2-10T (98.62%). The neighbor-joining tree constructed with 16S rRNA gene sequences of all valid Micromonospora species confirmed the taxonomic position of this strain that belongs to the genus Micromonospora and forms a monophyletic clade with M. auratinigra TT1-11T (Figure 2). Additionally, the maximum-likelihood tree also revealed that strain DCWR9-8-2T is closely related to M. auratinigra TT1-11T (Supplementary Figure S1). On basis of morphological, chemotaxonomic and phylogenetic data, this strain should be classified in the genus Micromonospora. Partial gyrB gene sequences of strain DCWR9-8-2T and other members in the genus Micromonospora were used for the multiple alignment and the sequence analysis. The partial gyrB gene sequence of strain DCWR9-8-2T displayed the highest sequence similarity (96.3%) with that of M. eburnea JCM12345T. Consistently, the phylogenetic analysis of the gyrB gene sequences showed that strain DCWR9-8-2T formed a cluster with M. eburnea JCM12345T. This was also significantly supported by the 68% bootstrap value (Supplementary Figure S3). The discrepancy between the phylogenetic relationship of strain DCWR9-8-2T based on the 16S rRNA and gyrB genes was similar to the results observed in previous studies. For example, the phylogenetic tree based on 16S rRNA gene sequences showed that M. tulbaghiae TVU1T was phylogenetically related to M. echinospora DSM 43816T and M. rosaria DSM 803T, while the phylogenetic analysis of the gyrB gene revealed that it was clustered together with M. aurantiaca NBRC 16155T and M. chalcea NBRC 13503T.38 Similarly, M. rhizosphaerae 211018T was closely related to M. olivasterospora DSM 43868T based on the phylogenetic analysis of the 16S rRNA gene sequences. However, the phylogenetic tree reconstructed from gyrB gene sequences indicated that its closest relative was M. inositola ATTC 21773T.39
Phylogenetic tree obtained by neighbor-joining tree based on almost-complete 16S rRNA gene sequences showing the position of strain DCWR9-8-2T among related species in the Micromonospora species. Catellatospora citrea subsp. Citrea IMSNU 22008T was used as outgroup. Asterisks (*) indicating the branches of the tree that were also found using the maximum-likelihood methods. The numbers on the branches indicate the percentage bootstrap values of 1000 replicates; only values >50% are indicated. Bar, 0.005 substitutions per nucleotide position.
Distinct chemotypic and phenotypic properties between strain DCWR9-8-2T and the closest phylogenetic relative, M. auratinigra TT1-11T are shown in Table 2. In particular, strain DCWR9-8-2T does not contain 3-OH-meso-diaminopimelic acid in its cell wall peptidoglycan while M. auratinigra TT1-11T presents this diaminopimelic acid. On the other hand, the presence of galactose in the cell hydrolysate is the significant point that distinguishes strain DCWR9-8-2T from M. auratinigra TT1-11T. The phenotypic data clearly discriminated strain DCWR9-8-2T from M. auratinigra TT1-11T as strain DCWR9-8-2T did not utilize d-fructose, d-melibiose, d-raffinose, d-ribose and salicin as the sole carbon sources, whereas M. auratinigra TT1-11T could utilize these carbon sources. Furthermore, the result of acid production from several carbon sources including the reduction of nitrate test also confirmed the differentiation between strain DCWR9-8-2T and M. auratinigra TT1-11T. Additionally, the low DNA–DNA relatedness value (32.1%±0.2 and 26.3%±0.8) was observed between strain DCWR9-8-2T, M. auratinigra TT1-11T and M. eburnea JCM12345T, respectively, below the 70% cut-off point suggested for the assignment of bacterial strains to the same genomic species.40 All above the differential characteristics confirmed that strain DCWR9-8-2T is a novel species of the genus Micromonospora, for which the name M. endophytica sp. nov. is proposed.
Description of Micromonospora endophytica sp. nov.
M. endophytica (en.do.phy’ti.ca. Gr. endo within; Gr. phyton plant; L. fem. suff. -ica adjectival suffix used with the sense of belonging to; N.L. fem. adj. endophytica within plant, endophytic, pertaining to the original isolation from plant tissue).
Gram-staining-positive, mesophilic actinobacteria that forms a single spore borne on the tip of the substrate mycelia. The orange yellow soluble pigment is observed on ISP2 and ISP3 media. Utilizes d-glucose, d-mannose, d-galactose, d-cellobiose, l-arabinose and d-xylose as sole carbon sources, weakly utilizes sucrose, but not glycerol, myo-inositol, l-rhamnose, d-ribose, d-fructose, d-raffinose, d-melibiose, mannitol, salicin and lactose. Acid production from l-arabinose and d-xylose. Peptonization of milk, hydrolysis of starch, gelatin liquefaction and nitrate reduction are positive. The maximum temperature for growth is 45 °C. The maximum NaCl concentration for growth is 2% (w/v). The pH range for growth is 5–10. The cell wall peptidoglycan contains meso-diaminopimelic acid. The acyl type of the cell wall is glycolyl. The predominant menaquinones is MK-9(H4) and MK-10(H6). The diagnostic whole-cell sugars are arabinose, glucose, ribose, xylose and small amount of mannose. The polar lipid profile contains diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, phosphatidylinositol, phosphatidylinositol mannosides, five unidentified phospholipids and two unidentified lipids but phosphatidylcholine is not detected. The major and sub-major fatty acids consists of iso-C16:0, iso-C15:0, anteiso-C17:0, anteiso-C15:0, iso-C17:0, C17:1 ω8c, 10-methyl C17:0, C18:1 ω9c, C17:0. Mycolic acids are absent. The G+C content of the DNA is 72.5 mol%. The type strain is DCWR9-8-2T (=BCC 67267T=NBRC 110008T), which was isolated from a leaf of upland rice collected from Chumporn province, Thailand
Accession code:
The DDBJ accession number for the 16S rRNA gene sequence of strain DCWR9-8-2T is AB981049.
Accession codes
References
Orskov, J. Investigations into the Morphology of the Ray Fungi, (Levin and Munksgaard, Copenhagen, (1923).
Schleifer, K. H. & Kandler, O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36, 407–477 (1972).
Weinstein, M. J. et al. Gentamicin, a New Antibiotic Complex from Micromonospora. J. Med. Chem. 6, 463–464 (1963).
He, H. et al. Lomaiviticins A and B, potent antitumor antibiotics from Micromonospora lomaivitiensis. Am. Chem. Soc 123, 5362–5363 (2001).
Igarashi, Y. et al. Lupinacidin C, an inhibitor of tumor cell invasion from Micromonospora lupine. J. Nat. Prod. 74, 862–865 (2011).
Igarashi, Y. et al. Maklamicin, an antibacterial polyketide from an endophytic Micromonospora sp. J. Nat. Prod. 74, 670–674 (2011).
Shirling, E. B. & Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340 (1966).
Itoh, T., Kudo, T., Parenti, F. & Seino, A. Amended description of the genus Kineosporia, based on chemotaxonomic and morphological studies. Int. J. Syst. Bacteriol. 39, 168–173 (1989).
Kelly, K. L. Inter-Society Color Council–National Bureau of Standard Color Name Charts Illustrated with Centroid Colors, (US Government Printing Office, Washington, DC, (1964).
Gordon, R. E., Barnett, D. A., Handerhan, J. E. & Pang, C. H. N. Nocardia coeliaca, Nocardia autotrophica, and the nocardin strain. Int. J. Syst. Bacteriol. 24, 54–63 (1974).
Arai, T. Culture Media for Actinomycetes, (The Society for Actinomycetes, Japan, Tokyo, (1975).
Williams, S. T. & Cross, T. Booth, C. Methods in Microbiology, (Academic Press, London Vol. 4, 295–334 (1971).
Kawamoto, I., Oka, T. & Nara, T. Cell wall composition of Micromonospora olivoasterospora, Micromonospora sagamiensis, and related organisms. J. Bacteriol. 146, 527–534 (1981).
Staneck, J. L. & Roberts, G. D. Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl. Microbiol. 28, 266–231 (1974).
Uchida, K. & Aida, K. An improved method for the glycolate test for simple identification of the acyl type of bacterial cell walls. J. Gen. Appl. Microbiol. 30, 131–134 (1984).
Komagata, K. & Suzuki, K. I. Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol. 19, 161–207 (1987).
Minnikin, D. E. et al. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 2, 233–241 (1984).
Sasser, M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids (MIDI Technical Note 101), MIDI: Newark, DE, (1990).
Kämpfer, P. & Kroppenstedt, R. M. Numerical analysis of fatty acid patterns of coryneform bacteria and related taxa. Can. J. Microbiol. 42, 989–1005 (1996).
Collins, M. D., Pirouz, T., Goodfellow, M. & Minnikin, D. E. Distribution of menaquinones in actinomycetes and corynebacteria. J. Gen. Microbiol. 100, 221–230 (1977).
Minnikin, D. E., Alshamaony, L. & Goodfellow, M. Differentiation of Mycobacterium, Nocardia, and related taxa by thin-layer chromatographic analysis of whole-organism methanolysates. J. Gen. Microbiol. 88, 200–204 (1975).
Tamaoka, J. Goodfellow, M. & O’Donnell., A. G. Chemical Methods in Prokaryotic Systematics 463–470 John Wiley & Sons: Chichester, (1994).
Tamaoka, J. & Komagata, K. Determination of DNA base composition by reversed-phase high-performance liquid chromatography. FEMS Microbiol. Lett. 25, 125–128 (1984).
Ezaki, T., Hashimoto, Y. & Yabuuchi, E. Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in micro-dilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int. J. Syst. Bacteriol. 39, 224–229 (1989).
Verlander, C. P. Kricka., L. J. Nonisotopic DNA Probe Techniques 185–201 Academic Press, New York, (1992).
Suriyachadkun, C. et al. Planotetraspora thailandica sp. nov., isolated from soil in Thailand. Int. J. Syst. Evol. Microbiol. 59, 992–997 (2009).
Garcia, L. C., Martínez-Molina, E. & Trujillo, M. E. Micromonospora pisi sp. nov., isolated from root nodules of Pisum sativum. Int. J. Syst. Evol. Microbiol 60, 331–337 (2010).
Lane, D. J. Stackebrandt, E. & Goodfellow, M. Nucleic Acid Techniques in Bacterial Systematics 115–148 John Wiley & Sons, Chichester, (1991).
Kim, O. S. et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721 (2012).
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic. Acids Res. 25, 4876–4882 (1997).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic. Acids Res. 22, 4673–4680 (1994).
Saitou, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Felsenstein, J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376 (1981).
Tamura, K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011).
Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120 (1980).
Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791 (1985).
Kawamoto, I. Williams, S. T., Sharpe, M. E. & Holt., J. G. Genus Micromonospora Orskov 1923, 147AL In Bergey’s Manual of Systematic Bacteriology, Williams & Wilkins, Baltimore Vol. 4, (2442–2450 (1989).
Kirby, B. M. & Meyers, P. R. Micromonospora tulbaghiae sp. nov., isolated from the leaves of wild garlic, Tulbaghia violacea. Int. J. Syst. Evol. Microbiol. 60, 1328–1333 (2010).
Wang, C. et al. Micromonospora rhizosphaerae sp. nov., isolated from mangrove rhizosphere soil. Int. J. Syst. Evol. Microbiol. 61, 320–324 (2011).
Wayne, L. G., Brenner, D. J. & Colwell, R. R. et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37, 463–464 (1987).
Acknowledgements
A research grant from King Mongkut’s Institute of Technology Ladkrabang Research Fund (KReF) is gratefully acknowledged. This study was supported in part by Department of Biology and Actinobacterial Research Unit, Faculty of Science, King Mongkut’s Institute of Technology, Ladkrabang, Bangkok, Thailand.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Rights and permissions
About this article
Cite this article
Thanaboripat, D., Thawai, C., Kittiwongwattana, C. et al. Micromonospora endophytica sp. nov., an endophytic actinobacteria of Thai upland rice (Oryza sativa). J Antibiot 68, 680–684 (2015). https://doi.org/10.1038/ja.2015.57
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2015.57
This article is cited by
-
Endophytic microbes: biodiversity, plant growth-promoting mechanisms and potential applications for agricultural sustainability
Antonie van Leeuwenhoek (2020)
-
Infection with Micromonospora strain SB3 promotes in vitro growth of Lolium multiflorum plantlets
Plant Cell, Tissue and Organ Culture (PCTOC) (2018)
-
Recent progress on the development of antibiotics from the genus Micromonospora
Biotechnology and Bioprocess Engineering (2016)