Abstract
Four new prenylated diphenyl ethers (1–4), diorcinols B-E, together with one known diorcinol (5), were obtained from the marine-derived fungus Aspergillus versicolor. Their structures were established on the basis of chemical and spectroscopic data. The absolute configurations of compounds 1 and 2 were determined by Mosher’s ester and specific rotation analysis. Their cytotoxicities were evaluated using A-549, Hela, BEL-7402 and K562 cell lines. Compound 3 exhibited moderate cytotoxicities against the Hela and K562 cell lines with the IC50 values of 31.5 and 48.9 μM, respectively, and compound 4 exhibited moderate cytotoxicity only against the Hela cell line with the IC50 value of 36.5 μM.
Similar content being viewed by others
Introduction
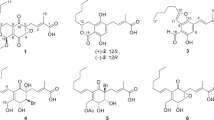
Marine-derived fungi belonging to the species Aspergillus versicolor are an attractive source of secondary metabolites with great structural diversities and interesting bioactivities, such as nitrobenzyl esters,1 chromones,2 sterigmatocystins3 and cyclopeptides.4 In our continuous search for new bioactive natural products from the marine-derived fungi, the fungus A. versicolor ZLN-60 was selected for the cyctoxicity in vitro against P388 cell line. In our previous report, two new cyclic pentapeptides, versicotides A and B, which showed no antitumor activity, were isolated from one of the fractions (Fr.3) of the ethyl acetate extract.5 Further investigation of the active fraction (Fr.2) led to the discovery of four new prenylated diphenyl ethers, diorcinols B-E (1–4) (Figure 1), together with a known compound, diorcinol (5).6 In this paper, we describe the isolation, structural elucidation and cytotoxic activities of these new compounds.
Results and discussion
The fungal strain was fermented and extracted as previously described.5 The ethyl acetate extract was concentrated in vacuo and then chromatographed by repeated silica column. The subfraction (Fr.2) with cytotoxic activity was further fractionated by Sephadex LH-20 and semipreparative HPLC to yield compounds 1–5 (Figure 1).
Structure determination
Diorcinol B (1) was obtained as yellow powder with a molecular formula of C19H24O5 deduced by the [M+H]+ ion peak at m/z 333.1714 in HR-ESI-MS spectrum, which indicated eight degrees of unsaturation. The 1H- and 13C-NMR spectra of 1 (Tables 1 and 2) displayed resonances that were assigned to four methyls, one methylene, one oxygen-bearing methane and five aromatic methines, five oxygen-bearing quaternary carbons (among them four aromatic carbons). These data suggested that 1 contains a diphenyl ether unit similar to diorcinol (5), which was also supported by the HMBC correlations.6 The major diffrences were the absence of one aromatic proton and the presence of an additional isopentyl chain unit in 1. The isopentyl group was determined by the 1H–1H COSY (H-1′′/H-2′′) and the HMBC correlations from H-4′′ (δH 1.62) and H-5′′ (δH 1.65) to C-2′′ (δC 78.7) and C-3′′ (δC 72.5). The HMBC correlations from H-1′′ (δH 2.63, 2.40) to C-1′ (δC 155.7), C-2′ (δC 121.7) and C-3′ (δC 140.4) located the isopentyl group on C-2′ (Figure 2). Considering the chemical shifts of C-2′′ and C-3′′, together with the molecular formula, the two hydroxyl groups were attached to C-2′′ and C-3′′.
The absolute configuration of C-2′′ was assigned as S by employing the modified Mosher’s method (Figure 3).7, 8
Diorcinol C (2), was separated as yellow powder, and an [M+H]+ molecular ion at m/z 347.1864 (calcd 347.1858) in the HR-ESI-MS spectrum was in agreement with the molecular formula C20H26O5. Their similar UV absorptions suggested that 2 was an analog of 1. On comparison of the 1D-NMR data for the two compounds (Tables 1 and 2), compound 2 showed an additional methoxy and the position was determined by the HMBC correlation between Me (3′′) (δH 3.08) and C (3′′) (δC 77.6) (Figure 2). Thus, compound 2 was elucidated as a methylated analog of morkeyol A (1). As the co-metabolite, 2 was proposed to have the same 2′′S-configuration, which was also supported by the similar specific rotation data (−29.3 in 2 versus −45.1 in 1).
Diorcinol D (3), was a yellow powder, and HR-ESI-MS indicated the molecular formula C19H22O3. Compound 3 displayed similar NMR resonances to morkeyol A (1) and their comparison suggested that they shared the same substructures of phenyl ethers (Tables 1 and 2). However, the absence of three signals for two hydroxy and one methenyl protons in 1 (δH 4.07, OH-2′′; δH 4.08, OH-3′′; and δH 3.35, H-2′′) and the presence of an additional olefinic singlet in 3 (δH 4.97, H-2′′) suggested that compound 3 contained an isopentenyl group. Therefore, the structure of 3 was determined by further analysis the HMBC correlations (Figure 2) and named as diorcinol D.
The molecular formula of 4 was determined as C19H22O4 on the basis of an [M+H]+ ion at m/z 315.1528 (calcd 315.1596) in the HR-ESI-MS. Similarities in the NMR data (Tables 1 and 2) of 4 and 3 showed that their structures were closely related. With the absence of the signals for the double bound and the appearance of an additional ketone (δc 211.8, C-2′′) and a methine (δc 40.4, C-3′′), together with the HMBC correlations from H-4′′ (δH 0.96) and H-5′′ (δH 0.97) to C-2′′ and C-3′′, and from H-1′′ (δH 3.68) to C-2′′ (Figure 2), the structure of 4 was deduced and named as diorcinol E.
Cytotoxic activities
The cytotoxicities of compounds 1–5 were evaluated in vitro on the A-549, BEL-7402, Hela and K562 cells by the SRB or 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazoliumbromide (MTT) methods.9, 10 Compound 3 exhibited moderate cytotoxicities against the Hela and K562 cell lines with the IC50 values of 31.5 and 48.9 μM, respectively, and compound 4 exhibited moderate cytotoxicity only against the Hela cell line with the IC50 value of 36.5 μM, whereas compounds 1, 2 and 5 showed no activities.
Discussion
A number of diphenyl ethers derivatives have been previously isolated and characterized from different organisms, whereas prenylated phenyl ethers are relatively rare in nature. To the best of our knowledge, only nine prenylated diphenyl ethers (tenellic acids) were isolated from several fungi such as Penicillium spp.11, 12 and Dendrospora tenella,13 and six natural transformation prenylated phenyl ethers products (chromenes) were found in the injured fruiting bodies of the mushrooms lactarius fuliginosus and lactarius picinus.14 These structurally similar diphenyl ethers showed diverse biological activities such as anti-microbial,11, 15, 16 cytotoxic17 and radical-scavenging activities.18 To our knowledge, this is the first report for the isolation of prenylated diphenyl ethers from a member of the genus Aspergillus, and compounds 3 and 4 exhibited cytotoxicities.
Materials and Methods
General
Specific rotations were obtained on a JASCO P-1020 digital polarimeter (JASCO, Tokyo, Japan). IR spectra were taken on a Nicolet Nexus 470 spectrophotometer (Thermo Scientific, Beijing, China) in KBr discs. NMR spectra were recorded on a JEOL JNM-ECP 600 spectrometer (JEOL Ltd, Beijing, China) using tetramethylsilane as internal standard, and chemical shifts were recorded as δ values. ESIMS utilized on a Q-TOF Ultima Global GAA076 LC mass spectrometer (Waters Asia, Ltd, Singapore). Semiprepartive HPLC was performed using an ODS column (HPLC:YMC-Pack ODS-A (5 μm, 10 × 250 mm, 4 ml min−1)). TLC and column chromatography were performed on plates precoated with silica gel GF254 (10–40 μm) and over silica gel (200–300 mesh, Qingdao Marine Chemical Factory, Qingdao, Shandong, China). Vacuum–liquid chromatography was carried out over silica gel H (Qingdao Marine Chemical Factory).
Fermentation and extraction of the fungal strain
The fungus strain was fermented and extracted as previously described.5
Purification
The crude extract (13.0 g) was applied to a silica gel (300–400 mesh) column and was separated into six fractions (Fr.1 to Fr.6) using a step-gradient elution of petroleum ether/CHCl3 and CHCl3/CH3OH. Fr.2 (1.28 g) was separated on a silica gel column using CHCl3 and second subfraction (Fr.2-2) further separated into four subfractions (Fr.2-2-1 to Fr.2-2-4) by Sephadex LH-20 (Greenherbs Science and Technology Development Co., Ltd., Beijing, China) using CH3OH as the eluting solvent. Subfraction Fr.2-2-1 was purified by semipreparative HPLC (50:50 MeOH/H2O, 4 ml min−1) to give compound 1 (15 mg, tR=9.5 min), 2 (13 mg, tR=13.0 min) and 4 (8 mg, tR=11.0 min). Subfraction Fr.2-2-3 was purified by semipreparative HPLC (60:50 MeOH/H2O, 4 ml min−1) to obtain compound 3 (19 mg, tR=15.0 min) and 5 (38.0 mg, tR=12.0 min).
The cytotoxic activities of subfractions were tested on 96-well plates and observed under a microscope (Supplementary Figure S1).
Physico-chemical properties
The MS, 1D and 2D NMR spectra of compounds 1–4 can be found in the Supplementary Figures S2–S25. Diorcinol B (1): yellow powder, [α]25D −45.1 (c 0.1, MeOH), UV (HPLC, mobile phase) λmax 210, 262 nm, IR (KBr) νmax 3206, 1588, 1467, 1320, 1144, 1064, 986 cm−1, 1H-NMR and 13C-NMR data (Tables 1 and 2), HR-ESI-MS m/z: 333.1714 [M+H]+ C19H25O5 (calcd 333.1702).
Diorcinol C (2): yellow powder, [α]25D −29.3 (c 0.1, MeOH), UV (HPLC, mobile phase) λmax 210, 262 nm, IR (KBr) νmax 3216, 1710, 1700, 1587, 1492, 1145, 1024, 1001 cm−1, 1H-NMR and 13C-NMR data (Tables 1 and 2), HR-ESI-MS m/z: 347.1864 [M+H]+ C20H27O5 (calcd 347.1858).
Diorcinol D (3): yellow powder, UV (HPLC, mobile phase) λmax 210, 262 nm, IR (KBr) νmax 3316, 1610, 1600, 1587, 1492, 1045, 1024, 985 cm−1, 1H-NMR and 13C-NMR data (Tables 1 and 2), HR-ESI-MS m/z: 299.1642 [M+H]+ C19H23O3 (calcd 299.1647).
Diorcinol E (4): yellow powder, UV (HPLC, mobile phase) λmax 210, 262 nm, IR (KBr) νmax 2964, 2917, 1710, 1700, 1587, 1467, 1322, 1105, 1024 cm−1, 1H-NMR and 13C-NMR data (see Tables 1 and 2), HR-ESI-MS m/z: 315.1582 [M+H]+ C19H23O4 (calcd 315.1596).
In vitro cytotoxicity assays
In the MTT assay, cell lines were grown in RPMI-1640 supplemented with 10% FBS in a humidified atmosphere of 5% CO2 and 95% air at 37 °C. Cell suspensions, 200 μl, at a density of 5 × 104 cells per ml were plated in 96-well microtiter plates and incubated for 24 h. Then, 2 μl of the test solutions (in MeOH) were added to each well and further incubated for 72 h. Then, 20 μl of the MTT solution (5 mg ml−1 in IPMI-1640 medium) was added to each well and incubated for 4 h. Old medium containing MTT (150 μl) was gently replaced by dimethylsulfoxide and pipetted to dissolve any formazan crystals formed. Absorbance was then determined on a Spectra Max Plus plate reader at 540 nm.
In the sulforhodanine B (SRB) assay, 200 μl of the cell suspensions were plated in 96-well plates at a density of 2 × 105 cells per ml. Then, 2 μl of the test solutions (in MeOH) was added to each well, and the culture was further incubated for 24 h. The cells were fixed with 12% trichloroacetic acid, and the cell layer was strained with 0.4% SRB. The absorbance of SRB solution was measured at 515 nm. Dose–response curves were generated, and the IC50 values, the concentration of compound required to inhibit cell proliferation by 50%, were calculated from the linear portion of log dose–response curves.
References
Belofsky, G. N., Jensen, P. R., Renner, M. K. & Fenical, W. New cytotoxic sesquiterpenoid nitrobenzoyl esters from a marine isolate of the fungus Aspergillus versicolor. Tetrahedron 54, 1715–1724 (1998).
Lin, W. H. et al. Novel chromone derivatives from the fungus Aspergillus versicolor isolated from the marine sponge Xestospongia exigua. J. Nat. Prod. 66, 57–61 (2003).
Lee, Y. M. et al. Bioactive metabolites from the sponge-derived fungus Aspergillus versicolor. Arch. Pharm. Res. 33, 231–235 (2010).
Zhuang, Y. B. et al. Cyclopeptides and polyketides from coral-associated fungus, Aspergillus versicolor LCJ-5-4. Tetrahedron 67, 7085–7089 (2011).
Zhou, L. N. et al. Two new cyclic pentapeptides from the marine-derived fungus Aspergillus versicolor. Helv. Chim. Acta 94, 1065–1070 (2011).
Itabashi, T., Nozawa, K., Nakajima, S. & Kawai, K. A new azaphilone, falconensin H, from Emericella falconensis. Chem. Pharm. Bull. 41, 2040–2041 (1993).
Dale, J. A. & Mosher, H. S. Nuclear magnetic resonance enantiomer regents. Configurational correlations via nuclear magnetic resonance chemical shifts of diastereomeric mandelate, O-methylmandelate, and α-methoxy-α-trifluoromethylphenylacetate (MTPA) esters. J. Am. Chem. Soc. 95, 512–519 (1973).
Ohtani, I., Kysumi, T., Kashman, Y. & Kakisawa, H. High-field FT NMR application of Mosher's method. The absolute configurations of marine terpenoids. J. Am. Chem. Soc. 113, 4092–4096 (1991).
Skehan, P. et al. New colorimetric cytotoxicity assay for anticancer-drug screening. J. Natl. Cancer Inst. 82, 1107–1112 (1990).
Mosmann, T. J. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 65, 55–63 (1983).
Zhang, Y. et al. Meroterpenoid and diphenyl ether derivatives from Penicillium sp. MA-37, a Fungus isolated from marine mangroverhizospheric soil. J. Nat. Prod. 75, 1888–1895 (2012).
Komai, S. et al. New penicillide derivatives isolated from Penicillium simplicissimum. J. Nat. Med. 60, 185–190 (2006).
Hyuncheol, O., Kwon, T. O., Gloer, J. B., Marvanova, L. & Shearer, C. A. Tenellic acids A-D: new bioactive diphenyl ether derivatives from the aquatic fungus Dendrospora tenella. J Nat. Prod. 62, 580–583 (1999).
Bernardi, M. D., Vidari, G. & Finzi, V. Fungal metabolites. 28. The chemistry of Lactarius fuliginosus and Lactarius picinus. Tetrahedron 48, 7331–7344 (1992).
Yamazaki, M. & Maebayashi, Y. Structure determination of violaceol-I and -II, new fungal metabolites from a strain of Emericella violacea. Chem. Pharm. Bull. 30, 514–518 (1982).
Bunyapaiboonasri, T., Yoiprommarat, S., Intereya, K. & Kocharin, K. New diphenyl ethers from the insect pathogenic fungus Cordyceps sp. BCC 1861. Chem. Pharm. Bull. 55, 304–307 (2007).
Nitao, J. K., Nair, M. G., Thorogood, D. L., Johnson, K. S. & Scriber, J. M. Bioactive neolignans from the leaves of Magnolia virginiana. Phytochemistry 30, 2193–2195 (1991).
Yang, G. H. et al. Induced production of halogenated diphenyl ethers from the marine-derived fungus Penicillium chrysogenum. Chem. Biodivers. 7, 2766–2770 (2010).
Acknowledgements
This work was financially supported by the National Natural Science Fundation of China (No. 41176120), the Promotive Research Fund for Excellent Young and Middle-aged Scientisits of Shandong Province (No. BS2010HZ027), the Program for New Century Excellent Talents in University (No. NCET-12-0499), the Public Projects of State Oceanic Administration (No. 2010418022-3) and the Program for Changjiang Scholars and Innovative Research Team in University (No. IRT0944).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Gao, H., Zhou, L., Cai, S. et al. Diorcinols B-E, new prenylated diphenyl ethers from the marine-derived fungus Aspergillus versicolor ZLN-60. J Antibiot 66, 539–542 (2013). https://doi.org/10.1038/ja.2013.40
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2013.40
Keywords
This article is cited by
-
Two New Diphenyl Ether Derivatives from the Fermentation Products of an Endophytic Fungus Phomopsis fukushii
Chemistry of Natural Compounds (2019)
-
Isopentylated diphenyl ether derivatives from the fermentation products of an endophytic fungus Phomopsis fukushii
The Journal of Antibiotics (2018)
-
New phenolic bisabolane sesquiterpenoid derivatives with cytotoxicity from Aspergillus tennesseensis
The Journal of Antibiotics (2018)
-
New Diorcinol J Produced by Co-Cultivation of Marine Fungi Aspergillus sulphureus and Isaria felina
Chemistry of Natural Compounds (2016)
-
Butyrolactones from the Fermentation Products of the Endophytic Fungus Aspergillus versicolor
Chemistry of Natural Compounds (2016)