Abstract
Actinomycetes were isolated from 109 soil and 93 leaf-litter samples collected at five sites in Vietnam between 2005 and 2008 using the rehydration-centrifugation (RC) method, sodium dodecyl sulfate-yeast extract dilution method, dry-heating method and oil-separation method in conjunction with humic acid-vitamin agar as an isolation medium. A total of 1882 strains were identified as Vietnamese (VN)-actinomycetes including 1080 (57%) streptomycetes (the genus Streptomyces isolates) and 802 (43%) non-streptomycetes. The 16S ribosomal RNA gene sequences of the VN-actinomycetes were analyzed using BLAST searches. The results showed that these isolates belonged to 53 genera distributed among 21 families. Approximately 90% of these strains were members of three families: Streptomycetaceae (1087 strains, 58%); Micromonosporaceae (516 strains, 27%); and Streptosporangiaceae (89 strains, 5%). Motile actinomycetes of the genera Actinoplanes, Kineosporia and Cryptosporangium, which have quite common morphological characteristics, were frequently isolated from leaf-litter samples using the RC method. It is possible that these three genera acquired common properties during a process of convergent evolution. By contrast, strains belonging to the suborder Streptosporangineae were exclusively isolated from soils. A comparison of the sampling sites revealed no significant difference in taxonomic diversity between these sites. Among the non-streptomycetes, 156 strains (19%) were considered as new taxa distributed into 21 genera belonging to 12 families. Interestingly, the isolation of actinomycetes from leaf-litter samples using the RC method proved to be the most efficient way to isolate new actinomycetes in Vietnam, especially the Micromonosporaceae species.
Similar content being viewed by others
Introduction
This is a study investigating the diversity and ecology of actinomycetes in Vietnam, and part of a joint research project between Vietnam and Japan. Vietnam is located in a tropical to subtropical region of Southeast Asia, from 8.3 to 22.3°N latitude, with 1700 km of coastline (north to south). The country has high geographical complexity ranging from mountainous land (500–1000 m above sea level) to watery lowland such as the Mekong Delta, hot springs and mangrove coasts. Climate and other ecological factors such as the availability of water, pH and organic contents of the soil affect the microbial flora. Additionally, there are 56 ethnic groups of people who eat many kinds of traditional fermented foods,1 thereby making the microbial gene pool more attractive. The presence of diverse and novel unique microbial species could be expected in the complex landscapes of Vietnam. Actinomycetes isolated in Vietnam are thought to be a potential source for screening for useful secondary metabolites.2, 3 A total of 1882 strains of actinomycetes isolated in Vietnam were included in a Vietnamese (VN)-actinomycetes collection. Publications comparing actinomycetic populations from different climates within Asia have been published. Xu et al.4 studied the diversity of soil actinomycetes in Yunnan (China), Wang et al.5 investigated the actinomycete diversity in the tropical rainforests of Singapore, Muramatsu et al.6 compared Malaysian and Japanese actinomycetes, and Ara and Kudo7, 8, 9 reported many novel genera of rare actinomycetes isolated from soil samples collected from Bangladeshi mangrove rhizospheres. Recently, Hayakawa et al.10 studied the diversity of actinomycetes isolated from soils in cool-temperate (Rishiri Island) and subtropical (Iriomote Island) areas of Japan. Here, we present results obtained from a complex study on ecology and taxonomy of actinomycetes isolated from soil and leaf-litter samples collected at five different sampling sites in Vietnam. The data on VN-actinomycetes is presented for the first time in this study and serves to enrich knowledge of the diversity and distribution of this microbial group in the region and the world.
Materials and methods
Sample collection
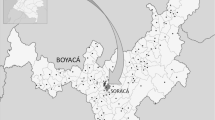
Between 2005 and 2008, 109 soil and 93 leaf-litter samples were collected from Vietnam, which is located in the tropical to subtropical regions of Indochina. The five sampling sites are shown in Figure 1. The diverse natural environment makes Vietnam an attractive country for a survey of novel microbial species including actinomycetes.
Isolation of actinomycetes
Four methods were used for the isolation of actinomycetes. The rehydration-centrifugation (RC) method11 was employed for isolating motile actinomycetes from soil and leaf-litter samples. Sodium dodecyl sulfate-yeast extract dilution method12 was used for general isolates from soil samples, while the dry-heating method13 allowed isolation of heat resistant strains from both soil and leaf-litter. The oil-separation (OS) method was used for lipophilic isolates from soil. In conjunction with these methods, humic acid-vitamin agar14 supplemented with nalidixic acid (20 mg l–1), cycloheximide (50 mg l–1) and kabicidin (20 mg l–1) was used as an isolation medium. All plates were incubated at 28–30 °C from 4 days to 3 weeks. Actinomycete colonies were picked and deposited on humic acid-vitamin agar, then purified by streaking onto yeast extract-starch agar (1% starch, 0.2% yeast extract and 2% agar, pH7.0). During these experiments, the biggest problem was the isolation of actinomycetes from environmental samples heavily contaminated with not only fungi and bacteria but also insects. Plates of isolates were sealed with parafilm and packaged into a plastic bag during cultivation.
The RC method used, one of the most important in this study, was modified to some degree from the original paper, with the method shown in Figure 2. Soil extract for sample suspension was prepared by suspending 500 g soil in 1 l of water, then autoclaved for 30 min and filtered. The OS method has been developed for selective isolation of lipophilic actinomycetes as described below. Approximately 0.5 g of dried soil samples were suspended in 5 ml of olive oil and mixed for 2 min. A 5 ml volume of sterilized water was added to the olive oil emulsion and mixed with a magnetic stirrer for 5 min, then centrifuged at 3000 r.p.m. (1500 g) for 10 min. The upper layer was diluted with fresh olive oil, and 0.1 ml of diluted samples were inoculated onto humic acid-vitamin agar and incubated at 28–30 °C for 1–3 weeks.
16S rRNA gene sequencing and phylogenetic analysis
Genomic DNA extraction was carried out using a Promega (Madison, WI, USA) extraction kit according to the manufacturer's protocol. The 16S ribosomal RNA (rRNA) gene was amplified by PCR using TaKaRa Ex Taq (Takara Bio, Otsu City, Shiga, Japan) with the primers, 9F (5′-GAGTTTGATCCTGGCTCAG-3′) and 1541R (5′-AAGGAGGTGATCCAGCC-3′), or occasionally 1510R (5′-GGCTACCTTGTTACGA-3′). Almost the entire sequence of the 16S rRNA gene (1300–1400 bp) was amplified by PCR as reported by Tamura and Hatano15 and directly sequenced using an ABI Prism BigDye Terminator cycle sequencing kit (Applied Biosystems, Foster City, CA, USA) and an ABI Model 3730 automatic DNA sequencer. The 16S rRNA gene sequence was compared with other sequences in the EMBL/GenBank/DDBJ database using BLAST searches and in the EzTaxon16 database, which includes only type strain sequences. The isolates demonstrating <98% identity compared with known species were considered as a potential novel species. Specifically, the 16S rRNA gene sequences obtained were aligned with reference sequences of known species in a genus using the MEGA ver. 5.01 soft package.17 A phylogenetic tree was constructed using neighbor-joining tree algorithms.18 The resultant neighbor-joining tree topology was evaluated by bootstrap analysis based on 1000 replicates.19
Results and Discussion
Isolation of actinomycetes from Vietnam
Between 2005 and 2008, 1882 strains were isolated in Vietnam, and were preserved in a VN-actinomycetes collection at the Institute of Microbiology and Biotechnology, Vietnam National University and the National Institute of Technology and Evaluation, Japan. These isolates were tentatively identified by analysis of the sequences of the 16S rRNA genes. Agar disks of actinomycete cultures packaged in a cryotube with 10% glycerol were kept at −80 °C for long-term preservation.
As shown in Table 1, the VN-actinomycetes collection was composed of 1080 streptomycetes (the genus Streptomyces strains) (57%) and 802 non-streptomycetes (43%). Streptomycetes are widely distributed throughout diverse natural environments. Since almost all streptomycetes grow quickly under conventional culture conditions, they are easily isolated. By contrast, non-streptomycetes, or rare actinomycetes, are generally characterized by slow growth and small colony formation. They are therefore difficult to isolate and to cultivate, especially in liquid media, so special isolation methods are required. The ratio of streptomycetes varies considerably each year; for example, 69% (348 strains) in 2005 and 43% (212 strains) in 2008. These percentages depend on isolation sources and isolation methods. In general, leaf-litter samples and the RC-method are relatively suited for non-streptomycetes isolation. The ratio of streptomycetes to non-streptomycetes is also influenced by unnatural factors such as the isolator's protocols. Therefore, we have mainly discussed the non-streptomycetes isolated from Vietnam.
The numbers of strains isolated from diverse samples and using various isolation methods are shown in Table 1. These isolates were composed of 1259 strains (67%) from soil and 623 strains (33%) from leaf-litter samples. Sampling was conducted by collecting the same numbers of soil and leaf-litter samples, respectively, from each sampling site. As the sodium dodecyl sulfate-yeast extract dilution method and the OS method were applied only for soil samples, and not with leaf-litter samples, the numbers of soil isolates were more numerous. The collection of VN-actinomycetes contained 706 strains (38%) using the RC method, 511 strains (27%) by the sodium dodecyl sulfate-yeast extract dilution method, 426 strains (23%) by the dry-heating method and 239 strains (13%) by the OS method. Owing to a technical problem in 2006, only 63 strains were obtained using the RC method (Table 1).
Taxonomic diversity of VN-actinomycetes
The generic identification of streptomycetes (1080 strains) was performed by observing their colony appearance and microscopic morphology, or by analysis of partial sequences of their 16S rRNA gene (9F, about 500 bp). In the case of all non-streptomycetes (802 strains), nearly the full length of the 16S rRNA gene sequences was determined and compared with known species in public databases, and their taxonomic positions were confirmed by phylogenetic analyses. As of December 2009, the term ‘Actinomycetes’ (order Actinomycetales) consists of 13 suborders, 42 families and about 200 genera based on the 16S rRNA gene sequence.20, 21 In this study, among the non-streptomycetes, 95 strains that were initially identified as members of the genera Actinoplanes, Catellatospora, Cellulomonas, Couchioplanes, Isoptericola or Micromonospora through BLAST searches of 16S rRNA gene sequence similarity were found to in fact belong to other genera through detailed phylogenetic analyses.
Family-level diversity
As shown in Table 2, VN-actinomycetes (1882 strains) were found to belong to 53 genera distributed among 21 families. At the family level, 58% (1087 strains) of the strains belonged to the family Streptomycetaceae. The most dominant group of non-streptomycetes belonged to the family Micromonosporaceae, in which there were 516 strains (27% VN-actinomycetes, 64% non-streptomycetes). The second dominant group (89 strains) of non-streptomycetes belonged to the family Streptosporangiaceae. The three families, Streptomycetaceae, Micromonosporaceae and Streptosporangiaceae, accounted for approximately 90% of strains isolated in the present study. Members of the families Kineosporiaceae, Pseudonocardiaceae and Cryptosporangiaceae were less frequently isolated.
Genus-level diversity
As mentioned already, sample types and isolation methods have been found to be well correlated with the taxonomic diversity of VN-actinomycetes (Table 2). As various actinomycetes belonging to the genera Actinoplanes, Kineosporia22, 23, 24 and Cryptosporangium15, 25 were frequently isolated from leaf-litter samples, as evidenced also by other reports,26, 27 it is conceivable that they may have an important role in the degradation of fallen leaves. It should be noted that these three genera belong to different families (Micromonosporaceae, Kineosporiaceae and Cryptosporangiaceae, respectively), which in turn belong to different suborders (Micromonosporineae, Kineosporiineae and Frankineae, respectively). This indicates that the actinomycetes we have isolated from leaf-litter samples are phylogenetically only remotely related with each other. Nonetheless, they share their habitats and show very similar characteristics such as possession of motility, absence or rarity of hydrophobic aerial hyphae, and formation of orange colonies, similar to the color of fallen leaves as shown in Figure 3a. As the moisture within fallen leaf deposits increases at the lower layers, motility and filamentous growth by substrate mycelium are potentially advantageous for the proliferation of actinomycetes within fallen leaf deposits. In our preliminary experiment, similar actinomycetes could not be isolated from fresh leaves or fresh fallen leaves, despite being frequently isolated from decomposed leaves. Therefore, it seems quite likely that the common characteristics possessed by the actinomycetes belonging to the three taxonomically distant genera mentioned above may have been independently acquired during the course of evolution.
Colony appearances of actinomycete isolates on various agar media. The strains on plates (a) were (in a clockwise direction from the top): 1, 2 and 3 Actinoplanes spp. (AB607853*, AB607849 and AB607850); 4 and 5 Kineosporia spp. (AB607851 and AB607854); and 6 Cryptosporangium sp. (AB607852) isolated from fallen leaves. Note the filmy roll back colonies of strains 1, 2 and 3 on ISP-2 medium (arrows). The strains on plates (b) were: 1 Pseudonocardia babensis VN05-A0561T; 2 Streptomyces sp. VN07-A0015; 3 New genus candidate (2) VN08-A0300; 4 New genus candidate (1) VN08-A0400; and 5 Kineosporia babensis VN05-A0415T. The agar media (from left to right) were yeast extract-starch agar, American Type Culture Collection (ATCC) medium-172 and ISP-2. The isolates were incubated at 28 °C for 10 days. *The DDBJ accession number on base sequences of 16S ribosomal RNA (rRNA) gene.
It should also be noted that, with the actinomycetes isolated from leaf-litter samples, particular species of bacteria were frequently co-isolated, perhaps reflecting the formation of a symbiotic community in their natural habitats. To separate these partners on decomposing organic matter and to obtain the rare actinomycetes in pure culture, the membrane method28 using a 0.22 μm pore size filter proved to be an effective procedure. Further analysis including molecular taxonomy of the bacteria thus separated may provide detailed features of their relationships as well as additional evidence for the apparent convergent evolution of the actinomycetes belonging to three different genera.
Conversely, 111 of the 113 strains belonging to the families Streptosporangiaceae, Nocardiopsaceae and Thermomonosporaceae, within the suborder Streptosporangineae, which form aerial mycelium, were isolated from soil samples (Table 2). Accordingly, these actinomycetes may have roles in the decomposition and recycling of organic matter, which is generally more difficult to degrade compared with fallen leaves. As mentioned above, the most dominant actinomycetes in the environment vary depending on the stage of decomposition of organic matter. Marked actinomycete diversity will also be a consequence of adaptation to this complex degradation process. The genera Streptomyces, Micromonospora, Dactylosporangium and Pseudonocardia were isolated from both soil and leaf-litter samples. With respect to isolation methods, motile actinomycetes such as members of Actinoplanes, Kineosporia, Cryptosporangium and Catenuloplanes were efficiently isolated by the RC method. Non-motile actinomycetes, such as Streptomyces (156 strains) or Micromonospora (34 strains) are distributed widely in nature and were also isolated by this method. The reason that the OS method is suitable for the isolation of Streptomyces, Micromonospora and Nonomuraea may be because their spore surfaces are lipophilic. There was no significant difference in actinomycete populations among the sampling sites across the northern and southern regions of Vietnam. However, there was a tendency that strains of Promicromonospora were more common (10/12 strains) in the north, and strains of Nocardiopsis were more common (6/7 strains) in the south.
Muramatsu et al.6 observed some interesting differences between the distribution of Malaysian (at latitude 3°N) and Japanese (35°N) actinomycete isolates. As an example, the number of strains belonging to the genera Streptosporangium and Nonomuraea within the family Streptosporangiaceae were 136 and 4, respectively, for Japanese isolates, and 20 and 69 for Malaysian isolates. In the present study of VN (12 to 22°N) actinomycetes, 11 Streptosporangium strains and 49 Nonomuraea strains were isolated. Wang et al.5 isolated 50 Streptosporangium and 390 Nonomuraea from the tropical rainforests of Singapore (2°N). Of the Indonesian (2–8°S) isolates,29 13 strains were identified as belonging to the genus Streptosporangium and 118 strains were members of Nonomuraea. More recent data reported by Hayakawa et al.10 comparing actinomycetes isolated in cool-temperate (45°N) and subtropical areas (24°N) in Japan also demonstrated a similar tendency. Additionally, on Mikurajim Island (34°N), Japan, 86 Streptosporangium strains and three Nonomuraea strains were isolated.30 It is interesting to note that the ratio of Streptosporangium to Nonomuraea strains is strongly dependent on the climate and the environment of their habitats and, as shown in Figure 4, the boundary of this change occurs at around 30°N, which coincides with the Watase Line, a well-known biogeographical boundary across the Tokara Islands of south-western Japan. It may be that cross-boundary differences in the variety of animal and plant species affect the environment in which these actinomycetes exist.
Species-level diversity
All the 16S rRNA gene sequences in non-streptomycetes (802 strains) were analyzed for species-level identification, and determined by BLAST searches. The results revealed a number of interesting phenomena regarding diversity among VN-actinomycetes at the species level. As an example, some dominant clusters existed, including new species, and there were also some peculiar endemic species in tropical and subtropical areas. We are now preparing as a sequel to the present study a paper describing species-level diversity of VN-actinomycetes. In previous studies, two new species, Kineosporia babensis31 (strain VN05-A0415T=NBRC 104154T=VTCC-A-0961T) and Pseudonocardia babensis32 (strain VN05-A0561T=NBRC 105793T=VTCC-A-1757T) have been identified. The colony appearance of these two type strains is shown in Figure 3b.
New taxon candidates
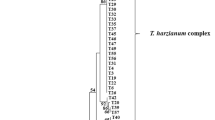
Actinomycete isolates demonstrating <98% identity of the 16S rRNA gene sequence to known species by BLAST searches and by EzTaxon server are generally considered to be new taxa.33, 34 As shown in Tables 3, 156 strains (19% of the non-streptomycetes) were regarded as new taxa, and were distributed into 21 genera in 12 families. One of the most important achievements of this study was the discovery that using the RC method in combination with leaf-litter samples proved to be the most reliable way to isolate new actinomycete species, especially new Micromonosporaceae species, in Vietnam. Many strains belonging to the family Micromonosporaceae (171 strains, 75%) were frequently considered to be new species, and mainly belonged to the genus Actinoplanes (95 strains, 61%). The use of the RC method is advantageous in order to isolate new species from leaf-litter samples. A total of 95 new species candidates were identified by detailed phylogenetic analyses; among these, 56 new species belonged to the genus Actinoplanes. Through phylogenetic analyses based on 16S rRNA gene sequences, three clades corresponding to a new genus have been found. These groups were designated as new genus candidates (1), (2) and (3) in Tables 2 and 3. As shown in Figure 5, the ‘new genus candidate (1)’ belongs to the family Microbacteriaceae, and the ‘new genus candidates (2) and (3)’ are members of the family Micromonosporaceae.
The new genus candidate (1) within the Microbacteriaceae was represented by strain VN08-A0400. This strain had 95.0% identity with other 16S rRNA gene sequences, the highest level among all the VN-actinomycetes. The morphology of strain VN08-A0400 was short rod-shaped (not filamentous), and the colony was black on yeast extract-starch agar and yellow on American Type Culture Collection medium 172 as shown in Figure 3b. Recently, Kim and Lee35 published a novel genus Amnibacterium within the family Microbacteriaceae, and strain VN08-A0400 seems to be closely related to this genus as shown in Figure 5.
On the basis of the 16S rRNA sequences, strain VN08-A0300 (new genus candidate (2)) exhibited 97.7% identity with strains of the genus Polymorphospora. Strain VN08-A0300 formed sporangia on dark orange colonies but lacked aerial hyphae (Figure 3b). As shown in Figure 5, strain VN07-A0427 and nine others strains belonged to new genus candidate (3) with 3–5 new species, forming a single clade closely related to the genus Krasilnikovia.
In conclusion, many novel actinomycetes have been discovered with high frequency in Vietnam, and are expected to be useful as a source of strains to be screened for production of novel secondary metabolites as well as for determining their new ecological roles in tropical and subtropical regions.
References
Thanh, V. N. Yeast, biodiversiy and promises. Proc. Natl Conf. Basic Biotechnol., Vietnam 118–123 (2005).
Bérdy, J. Bioactive microbial metabolites. J. Antibiot. 58, 1–26 (2005).
Sezaki, M. & Miyadoh, S. Practically used antibiotics and their related substrates. in Identification Manual of Actinomycetes (ed. Miyadoh, S.) 349–389 (Mainichi Academic Forum, Tokyo, 2002).
Xu, L. H., Li, Q. R. & Jiang, C. L. Diversity of soil actinomycetes in Yunnan, China. Appl. Environ. Microbiol. 62, 244–248 (1996).
Wang, Y., Zhang, J. S., Ruan, J. S., Wang, Y. M. & Ali, S. M. Inveatigation of actinomycete diversity in the tropical rainforests of Singapore. J. Ind. Microbiol. Biotechnol. 23, 178–187 (1999).
Muramatsu, H., Shahab, N., Tsurumi, Y. & Hino, M. A comparative study of Malaysian and Japanese actinomycetes using a simple identification method based on partial 16S rDNA sequence. Actinomycetologica 17, 33–43 (2003).
Ara, I. & Kudo, T. Krasilnikovia gen. nov., a new member of the family Micromonosporaceae and description of Krasilnikovia cinnamonea sp. nov. Actinomycetologica 21, 1–10 (2007).
Ara, I. & Kudo, T. Luedemannella gen. nov., a new member of the family Micromonosporaceae and description of Luedemannella helvata sp. nov. and Luedemannella flava sp. nov. J. Gen. Appl. Microbiol. 53, 39–51 (2007).
Ara, I. & Kudo, T. Sphaerosporangium gen. nov., a new member of the family. Streptosporangiaceae. Actinomycetologica 21, 11–21 (2007).
Hayakawa, M. et al. Diversity analysis of actinomycetes assemblages isolated from soils in cool-temperate and subtropical areas of Japan. Actinomycetologica 24, 1–11 (2010).
Hayakawa, M., Otoguro, M., Takeuchi, T., Yamazaki, T. & Iimura, Y. Application of a method incorporating differential centrifugation for selective isolation of motile actinomycetes in soil and plant litter. Antonie Van Leeuwenhoek 78, 171–185 (2000).
Hayakawa, M. & Nonomura, H. A new method for the intensive isolation of actinomycetes from soil. Actinomycetologica 3, 95–104 (1989).
Hayakawa, M., Sadataka, T., Kajiura, T. & Nonomura, H. New methods for the highly selective isolation of Micromonospora and Microbispora from soil. J. Fermen. Bioeng. 72, 320–326 (1991).
Hayakawa, M. & Nonomura, H. Humic acid-vitamin agar, a new medium for the selective isolation of soil actinomycetes. J. Ferment. Technol. 65, 501–509 (1987).
Tamura, T. & Hatano, K. Phylogenetic analysis of the genus Actinoplanes and transfer of Actinoplanes minutisporangius Ruan et al. 1986 and ‘Actinoplanes aurantiacus’ to Cryptosporangium minutisporangium comb. nov. and Cryptosporangium aurantiacum sp. nov. Int. J. Syst. Evol. Microbiol. 51, 2119–2125 (2001).
Chun, J. et al. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences (www.eztaxon.org). Int. J. Syst. Evol. Microbiol. 57, 2259–2261 (2007).
Tamura, K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. and Evol. (in press).
Saitou, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791 (1985).
Zhi, X. Y., Li, W. J. & Stackebrandt, E. An update of the structure and 16S rRNA gene sequence-based definition of higher ranks of the class Actinobacteria. Int. J. Syst. Evol. Microbiol. 59, 589–608 (2009).
Euzéby, J. P. List of Procaryoteic names with standing in nomenclature. (http://www.bacterio.cict.fr/index.html).
Pagani, H. & Parenti, F. Kineosporia, a new genus of the order Actinomycetales. Int. J. Syst. Bacteriol. 28, 401–406 (1978).
Itoh, T., Kudo, T., Parenti, F. & Seino, A. Amended description of the genus Kineosporia, based on chemotaxonomic and morphological studies. Int. J. Syst. Bacteriol. 39, 168–173 (1989).
Kudo, T., Matsushima, K., Itoh, T., Sasaki, J. & Suzuki, K. Description of four new species of the genus Kineosporia: Kineosporia succinea sp. nov., Kineosporia rhizophila sp. nov., Kineosporia mikuniensis sp. nov. and Kineosporia rhamnosa sp. nov., isolated from plant samples, and amended description of the genus Kineosporia. Int. J. Syst. Bacteriol. 48, 1245–1255 (1998).
Tamura, T., Hayakawa, M. & Hatano, K. A new genus of the order Actinomycetales, Cryptosporangium gen. nov., with descriptions of Cryptosporangium arvum sp. nov. and Cryptosporangium japonicum sp. nov. Int. J. Syst. Bacteriol. 48, 995–1005 (1998).
Makkar, N. S. & Cross, T. Actinoplanes in soil and on plant-litter from freshwater habitats. J. Appl. Bacteriol. 52, 209–218 (1982).
Willoughby, L. G. A study on aquatic actinomycetes, the allochthonous leaf component. Nova Hedwigia 18, 45–113 (1969).
Hirsch, C. F. & Christensen, D. L. Novel method for selective isolation of actinomycetes. Appl. Environ. Microbiol. 46, 925–929 (1983).
Lisdiyanti, P. & Otoguro, M. The number of isolates based on genera of Indonesian actinomycetes. in Taxonomic and Ecological Studies of Fungi and Actinomycetes in Indonesia (eds Windyasturi, Y. & Ando, K.) 601–602 (2010).
Ohno, M. & Harayama, S. Isolation of actinomycetes from Japanese soil. in Explaration of Microorganisms in Diverse Environments (article in Japanese, ed. Harayama, S.) 154–162 (2008).
Sakiyama, Y. et al. Kineosporia babensis sp. nov., isolated from plant litter in Vietnam. Int. J. Syst. Evol. Microbiol. 59, 550–554 (2009).
Sakiyama, Y. et al. Pseudonocardia babensis sp. nov., isolated from plant litter in Vietnam. Int. J. Syst. Evol. Microbiol. 60, 2336–2340 (2010).
Drancourt, M. et al. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J. Clin. Microbiol. 38, 3623–3630 (2006).
Stackebrandt, E. & Ebers, J. Taxonomic parameters revisited: tarnished gold standards. Microbiol. Today 33, 152–155 (2006).
Kim, S. J. & Lee, S. S. Amnibacterium kyonggiense gen. nov., sp. nov., a novel genus of the family Microbaceriaceae. Int. J. Syst. Evol. Microbiol. 61, 155–159 (2011).
Acknowledgements
This study was founded and conducted as a joint research project between the Institute of Microbiology and Biotechnology, Vietnam National University, Hanoi, Vietnam (VNUH-IMBT), and the Biological Resource Center, National Institute of Technology and Evaluation (NBRC), Japan. We thank Mr and Mrs Lechevalier, Drs S Ikeda, M Hayakawa, H Muramatsu, K Isono, I Okane, T Kuzuyama, T Nakashima, T Tamura, P Hoa, P Lisdiyanti and Sumi for their useful discussion and valuable comments on the paper.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Van Hop, D., Sakiyama, Y., Binh, C. et al. Taxonomic and ecological studies of actinomycetes from Vietnam: isolation and genus-level diversity. J Antibiot 64, 599–606 (2011). https://doi.org/10.1038/ja.2011.40
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2011.40
Keywords
This article is cited by
-
Development of a drug discovery approach from microbes with a special focus on isolation sources and taxonomy
The Journal of Antibiotics (2023)
-
Multiple strain analysis of Streptomyces species from Philippine marine sediments reveals intraspecies heterogeneity in antibiotic activities
Scientific Reports (2021)
-
Antimicrobial biosynthetic potential and diversity of culturable soil actinobacteria from forest ecosystems of Northeast India
Scientific Reports (2020)
-
Virgisporangium myanmarense sp. nov., a novel motile actinomycete isolated from an anthill soil in Myanmar
The Journal of Antibiotics (2017)
-
Antimicrobial activities of actinomycetes isolated from unexplored regions of Sundarbans mangrove ecosystem
BMC Microbiology (2015)