Abstract
Monensin A phenylurethane sodium salt (MON-UR1-Na) crystals were studied by the X-ray, NMR, FT-IR and PM5 semi-empirical methods. The X-ray data show that the compound forms a pseudocyclic structure, stabilized by three intramolecular hydrogen bonds, and the sodium cation coordinated by five oxygen atoms in the hydrophilic sphere. The NMR and FT-IR data demonstrate that this pseudocyclic structure is also conserved in CH2Cl2 solution. This structure of MON-UR1-Na is significantly different than the ones previously proposed by Westley et al. and Tanaka et al. The semi-empirical calculations of the MON-UR1-Na structures indicate that the one of the crystal is the most energetically favorable one. Other parameters, such as the size, chemical and biological nature of the urethane substituent, and especially the free carbonyl urethane group, may have a role in the biological activity of MON-UR1-Na. The in vitro microbiological tests provide evidence that MON-UR1-Na shows higher antibacterial activity against human pathogenic bacteria, including antibiotic-resistant Staphylococcus aureus and Staphylococcus epidermidis than the parent unmodified antibiotic—Monensin A.
Similar content being viewed by others
Introduction
Semi-synthetic and synthetic urethanes (carbamates) have been subjects of intensive studies because of their applications in the agrochemical industry as herbicides, fungicides and in medicine as pharmaceuticals. They can be used as anticancer, antimicrobial and antimalarial agents, and in many other areas.1, 2 The chemistry of urethanes as anticancer agents has mainly been used to develop the so-called prodrugs. It was also shown that the presence of one urethane residue in the compounds demonstrating biological activity improves their pharmacological and pharmacokinetic properties.2
Several research groups have investigated the influence of urethane residues in the respective derivatives of monensin A on their antibacterial, anticoccidial, antiparasitic and antimalarial activity (Scheme 1).3, 4, 5, 6, 7, 8, 9 It has been also shown that the series of urethane derivatives of monensin A function as growth agents in ruminants. Furthermore, these compounds have also antihypertensive and antimalarial activity, and were used in the treatment of swine dysentery.8, 9
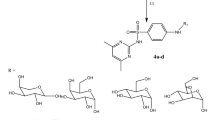
Monensin A (Scheme 1) isolated from Streptomyces cinnamonensis is a typical representative of natural polyether ionophore antibiotics, capable of forming pseudomacrocyclic complexes with metal cations.10, 11, 12, 13, 14, 15, 16, 17, 18 The antimicrobial properties of monensin result from its ability to transport metal cations through cellular and subcellular membranes and disrupt the Na+/K+ ion balance, which finally leads to the cell death.19, 20, 21, 22 Actually, monensin A sodium salt (MON-Na) is used in veterinary medicine as a growth-promoting agent and as a coccidiostatic agent.
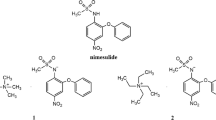
Westley et al.3, 4, 9 reported that several phenyl urethanes of monensin A exhibit higher antibacterial activity than pure monensin. Literature provides several possible explanations of the differences in biological activity between the (C-26) urethane derivatives of monensin A and the parent antibiotic.3, 4, 5, 6, 7, 8 The study of monensin A has shown that the C(26)OH group is essential for the remarkable stability of the 1:1 sodium complex in the natural ionophore.3, 4 Chemical modifications of the C(26)OH moiety in monensin A affect the Na+/K+ selectivity and change its antibiotic activity.5, 6, 7 When the C(26)OH hydroxyl group is transformed to an urethane group, the efficiency of Na+ transport through the membrane decreases, whereas that of K+ transport increases.5, 6 A further explanation is that, in contrast with monensin, the urethanes have the ability to transport divalent metal cations. All these explications are linked to the structure of the monensin A urethane complex with sodium cation.4 Up till now, two different structures of such a complex have been proposed (Scheme 2).3, 4, 7 The first structure of the monensin-urethane sodium salt has been proposed by Westley et al.3, 4 In this structure, the carbonyl oxygen atom of urethane group is engaged in the coordination process of the sodium cation (Scheme 2a), enhancing the stability of this complex. An alternative structure of the monensin urethane sodium salt complex has been proposed by Tanaka et al.,7 who postulated a head-to-tail pseudocyclic structure of the sodium complex stabilized by two intramolecular hydrogen bonds. One hydrogen bond is formed between N-H proton from the urethane group and the O(2) oxygen atom of the carboxylate group, and a second one is formed between the C(25)O-H proton and the O(1) oxygen atom of the carboxylate group (Scheme 2b).
In this paper, C(26)-O-phenylurethane of MON-Na (MON-UR1-Na) was synthesized and studied in the solid state by X-ray crystallography, and in solution by NMR and FT-IR spectroscopic methods. The structure of MON-UR1-Na was also visualized by the PM5 calculations.
Preliminary tests of the antimicrobial activity of MON-UR1-Na showed that it was active against Gram-positive standard bacterial strains.5, 7 This result inspired us to test the antibacterial activity of MON-UR1-Na on a series of clinical isolates of methicillin-resistant Staphylococcus aureus and S. epidermidis. Staphylococci are very troublesome pathogens in hospitals and other healthcare settings. They represent a group of invasive, biofilm-forming microorganisms responsible for the nosocomial infections. Increasing resistance of staphylococci to antibiotics and other antimicrobials is the reason for widespread search for new antimicrobial agents.23, 24, 25
Results and discussion
Chemistry
The structures of monensin A (MONA), MON-Na and C(26)-O-MON-UR1-Na, together with the atom numbering are shown in Scheme 1. The conversion of MON-Na to its phenylurethane was carried under mild condition in a simple reaction between MON-Na and phenyl isocyanate in CH2Cl2 as a solvent. This one-pot reaction led to MON-UR1-Na with a good yield (65%). The product was easily purified by column chromatography. The structure of the product was determined using the elemental analysis and the X-ray, FT-IR, 1H and 13C NMR methods, and is discussed in detail below.
Crystal and molecular structures of MON-UR1-Na
The crystal structure of MON-UR1-Na at 293 K was solved using single-crystal X-ray diffraction (Tables 1, 2 and 3, and Supplementary Tables S1 and S2). The crystal is built of globular complex molecules with relatively strong hydrogen bonds formed within them. The anion is wrapped around the sodium cation, and the cation is fivefold coordinated by three THF, one hydroxy and one carboxylate oxygen atom (Figure 1, Table 2). The length of Na–O coordination bonds varies from 2.315(4) Å for THF O(8) oxygen atom to 2.547(4) Å for THF O(7) oxygen atom. There are three intramolecular hydrogen bonds binding the terminal parts of the MON-UR1 anion, listed in Table 3. In the crystal, there are no short intermolecular contacts, except for van der Waals ones (Supplementary Figure S1). The coordination in the MON-UR1-Na and MON-Na molecules is very similar (Tables 2, 3 and 4, Figure 2),15 except the etheric O(9) and hydroxyl O(10) oxygen atom exchange in coordinating Na because of the rotation of the tetrahydropyran ring.
FT-IR studies
In Figure 3a, the FT-IR spectra of crystalline MON-UR1-Na in nujol/fluorolube mulls (solid line) and in CH2Cl2 solution (dashed line) are compared. The same spectra with extended scales in the regions of the ν(OH) and ν(NH), as well as ν(C=O) vibrations are shown in Figures 3b and c, respectively.
According to the crystallographic data of MON-UR1-Na, the O(4)H, O(10)H and N(1)H protons are involved in the intramolecular hydrogen bonds with the carboxylate anion (Table 3). Two of these hydrogen bonds are clearly stronger than the one O(4)-H⋯O(2). This situation leads to the formation of a complex band with maxima at 3340 cm−1, 3238 cm−1, 3188 cm−1, 3130 cm−1 assigned to protonic vibrations in these three different intramolecular hydrogen bonds existing within the crystal structure. The parameters of those hydrogen bonds are shown in Table 3.
Secondary urethanes absorb near 3450–3390 cm−1 because of the N-H stretching vibration if hydrogen bonding is absent and about 3340 cm−1 if the N-H proton is involved in the hydrogen bond.26 In the spectrum of MON-UR1-Na, no band in the 3450–3390 cm−1 range is observed, indicating that the N-H proton is hydrogen bonded.
In the FT-IR spectrum of crystalline MON-UR1-Na, the ν(C=O) (amide I) band of urethane and the νas(COO–) band of the carboxylate group are observed at 1735 and 1572 cm−1, respectively, (Figure 3c). The νas(COO–) band is relatively broad because it overlaps with the amide II band of the urethane group.
A comparison of the FT-IR spectra of MON-UR1-Na in the crystal and the CH2Cl2 solution shows only small changes in the shape of the band complex in the region over 3050 cm−1 (Figure 3b), indicating that all the intramolecular hydrogen bonds and the structure observed in the solid are conserved also in the solution. Only small spectral changes are observed in the region between 1800 cm−1 and 1500 cm−1 (Figure 3c). In this region, the MON-UR1-Na spectrum in the solid state (solid line) exhibits only one band at 1735 cm−1 assigned to the ν(C=O) vibrations of C(37)=O group. In the spectrum in solution, this band becomes broadened and is slightly shifted to 1728 cm−1. Also the amide II band becomes more pronounced showing a maximum at 1557 cm−1. These spectral differences strongly suggest that in the solution the electron distribution within the urethane group is slightly different than in the solid, that is, the C(37)=O group in the solution becomes slightly more polar and the N-H proton becomes slightly stronger hydrogen bonded with the carboxylate group. The last observation can also explain the vanishing of the shoulder, observed at 3340 cm−1 in the spectrum of the solid and assigned to the ν(N-H) vibrations, in the spectrum in the solution. It is also in agreement with the previous spectroscopic studies of various urethanes.27
NMR studies
The 1H- and 13C NMR data of MON-UR1-Na in CD2Cl2 are summarized in Table 5. The 1H- and 13C NMR signals were assigned using one- and two-dimensional such as COSY, HETCOR, NOESY, HMBC spectra shown in Supplementary Figures S2–S5, respectively, (in the Supplementary Materials) as well as by the addition of CD3OD to the sample (Figure 4). In the 1H NMR spectrum of MON-UR1-Na, the signals of the two OH and one NH protons are separate (Figure 4). The signals observed at 3.81 and 7.81 p.p.m., are assigned to the O(4)H and O(10)H protons, respectively, and the N(1)H proton signal is observed at 9.62 p.p.m.. The chemical shifts of these signals indicate that this O(4)H group is involved in a very weak hydrogen bond and O(10)H and especially the N(1)H groups are involved in much stronger hydrogen bonds, which is in agreement with the X-ray data discussed previously (Table 3) as well as with the FT-IR data discussed above.
The crystallographic data show that coordination of the sodium cation is realized in a pseudocyclic structure of the MON-UR1-Na molecule. The existence of this head-to-tail pseudocyclic structure of MON-UR1-Na in the solution is confirmed by the NMR NOESY spectrum in which the coupling between the O(4)H and O(10)H proton is observed, indicating clearly that these OH groups should be relatively close in space. Thus, cyclic conformation present in the crystal is conserved in the solution.
Semi-empirical calculations
On the basis of the spectroscopic as well as X-ray results, the structure of MON-UR-1 has been calculated by the MM3 molecular mechanics and PM5 semi-empirical method and compared with the crystal structure (Figure 5). The structure implied by the semi-empirical calculations is similar to that obtained by the X-ray diffraction data. The only difference between the crystallographic data and the respective calculations result from the fact that the data compared have been obtained for two different phases: solid and gas, and from the lack of interactions between the molecules of MON-UR1-Na.
The structures of MON-UR1-Na proposed by Westley et al.3, 4 as well as by Tanaka et al.7 (Figures 5b and c) have also been calculated. The calculated heats of formation of these structures indicate that the most stable complex in the gas phase has the structure proposed by us in this paper, based on X-ray and spectroscopic studies (heats of formation=−642.3 kcal mol−1). The calculated heats of formation values for structures proposed by Westley and Tanaka are −614.5 kcal mol−1 and −603.2 kcal mol−1, respectively, demonstrating that these structures are energetically unfavorable. This result is in good agreement with the spectroscopic and crystallographic data discussed above.
Antimicrobial activity
Preliminary tests of the antimicrobial activity of several urethanes of monensin showed that it was active against Gram-positive standard bacterial strains. It has been shown that several urethanes of monensin exhibited higher antibacterial activity than pure monensin.3, 4, 9 This result inspired us to verify in vitro the antibacterial activity of MON-UR1-Na on typical Gram-positive cocci, Gram-negative rods and yeast-like organisms, as well as on a series of clinical isolates of Staphylococcus: methicillin-resistant S. aureus and methicillin-resistant S. epidermidis. Hospital strains of methicillin-resistant Staphylococcus were isolated from different biological materials of patients of the Warsaw Medical University Hospital. Unfortunately, infections caused by this organism are becoming more difficult to treat as further evolution of drug resistance occurs within the pathogen.22, 23, 24, 25, 28, 29
The data regarding the antimicrobial activity of the compounds are summarized in Tables 6 and 7. MON-Na and MON-UR1-Na are very active against Gram-positive bacteria. Phenyl urethane of monensin shows higher antibacterial activity (MIC=0.25–4 μg ml−1) in comparison with that of MON-Na (MIC=2.0–12.5 μg ml−1). It is interesting to note that MON-UR1-Na exhibits relatively low antifungal activity (MIC=100 μg ml−1) against Candida, whereas MON-Na is inactive against these fungi. Moreover, as expected, the cell walls of Gram-negative bacteria do not permit the penetration of hydrophobic molecules with high weights, and thus the microorganisms are not susceptible to the action of monensin. The same theory applies to its urethane.
The antimicrobial activity results given in Table 7 demonstrate that MON-UR1-Na shows higher antistaphylococcal activity in comparison with that of MON-Na as well as higher in comparison with that of monensin esters and amides.30, 31, 32, 33, 34 For most isolates tested, the MIC values were about 0.5 μg ml−1. Only four strains of MRSE and one of MRSE among the strains tested were slightly more resistant to MON-UR1-Na, reaching a MIC value of about 1 μg ml−1.
We provide evidence that phenyl urethane of monensin shows higher antibacterial activity against human pathogenic bacteria, including antibiotic-resistant S. aureus and S. epidermidis, than the parent unmodified antibiotic. The results suggest that such a modification of monensin direction is appropriate and could give potential drugs to combat staphylococcal infections.
The highest antibacterial activity of MON-UR1-Na as well as the slight antifungal activity of monensin urethane are probably related to the presence of the urethane moiety, as it was previously observed that various compounds containing this substituent show moderate antibacterial and strong antifungal activities.1, 2
Conclusions
The interactions between the etheric oxygen atoms of monensin and its phenyl urethane with sodium cation lead to the formation of the pseudocyclic structures, which are stabilized by different intramolecular hydrogen bonds (Figure 1, Table 3). By this investigation, we have proved that the oxygen atom of the C=O urethane group is not engaged in the coordination of the sodium cations as postulated by Westley et al.3, 4 The X-ray and spectroscopic studies show clearly that the system of intramolecular hydrogen bonds present in the molecular structure of MON-UR1-Na is also different to that proposed by Tanaka et al.7 Thus, differences in the biological activities between monensin and its urethane derivative described here are not based on a different capability of complex formation but on other parameters such as size and chemical and biological nature of the urethane substituent, and especially on the stability of the complex in which the carbonyl urethane group is not involved in the complexation process.
Experimental procedure
Preparation of C(26)-O-phenyl urethane of MON-UR1-Na
All precursors for the syntheses (MON-Na and phenyl isocyanate) and solvents were obtained from Aldrich or Fluka (Poznan, Poland) and were used as received without further purification. CD2Cl2 and CD2Cl2 spectral-grade solvents were stored over 3 Å molecular sieves for several days. All manipulations with the substances were performed in a carefully dried and CO2-free glove box. TLC was carried out on precoated plates (TLC silica gel 60 F254, Aluminum Plates Merck, Darmstadt, Germany), and spots were detected by illumination with an UV lamp and visualized with iodine or by spraying 1% Ce(SO4)2-10% H2SO4, followed by heating. All the solvent used in flash chromatography was of HPLC grade (CHROMASOLV from Sigma-Aldrich, Poznan, Poland) and were used as received. The elemental analysis of MON-UR1-Na was carried out on Vario ELIII (Elementar, Hanau, Germany).
Synthesis
MON-Na (1 g, 1.49 mmol) and phenyl isocyanate (215 mg, 1.8 mmol) were dissolved in dry CH2Cl2 (30 ml) and stirred at room temperature. The course of the reaction was followed by silica gel TLC. After 2 weeks, the reaction appeared complete and the reaction mixture was evaporated to dryness under reduced pressure and purified using dry-column flash chromatography35, 36 (Silica gel 60 from Fluka), first eluted with CH2Cl2 to separate the traces of phenyl isocyanate and then with the CH2Cl2/CH3OH (from 100:1 to 10:1) solvent mixture as an eluent, to give MON-UR1-Na (65% yield). The MON-UR1-Na was crystallized from 1:1 mixture of CH2Cl2 and hexane. Elemental analysis: calculated for C43H66NNaO12: C, 63.61%; H, 8.19%; N, 1.73%. Found: 63.43%; H, 8.24%; and N, 1.55%.
X-ray measurements
The crystals selected for single-crystal X-ray diffraction measurements formed colorless parallelepipeds with well-developed faces. They were stable under normal conditions and the X-ray diffraction measurements were carried out on an Oxford Diffraction Supernova diffractometer (Oxford Diffraction, Abingdon, UK) using CuKα radiation at room temperature. The structure was solved by direct methods37 and refined by full-matrix least squares.38 One H-atom at O(4) was located from the difference Fourier maps and refined with isotropic temperature factor. All other H atoms were located from the molecular geometry (C-H 0.93 and 0.98, O-H 0.82, N-H 0.86 Å) and their Uiso's were related to the thermal vibrations of their carriers. Selected details about the crystal structure, experiment, structure solution and refinement are given in Table 1. The crystallographic information file has been deposited at the Cambridge Crystallographic Database Center as a supplementary publication no. CCDC 778665.
Spectroscopic measurements
The 1H, 13C and 15N NMR spectra were recorded on a Bruker Avance DRX 600 spectrometer (Bruker, Karlsruhe, Germany). 1H NMR measurements of MON-UR1-Na (0.07 mol dm−3) in CD2Cl2 were carried out at the operating frequency of 600.055 MHz; flip angle=45°; spectral width=4500 Hz; acquisition time=2.0 s; relaxation delay=1.0 s; T=293.0 K and using TMS as the internal standard. No window function or zero filling was used. Digital resolution was 0.2 Hz per point. The error of the chemical shift value was 0.01 p.p.m. The 13C NMR spectra were recorded at the operating frequency 150.899 MHz; flip angle=60°; spectral width=19000 Hz; acquisition time=1.8 s; relaxation delay=1.0 s; T=293.0 K and TMS as the internal standard. Line-broadening parameters were 0.5 or 1 Hz. The error of chemical shift value was 0.1 p.p.m. All spectra were locked to deuterium resonance of CD2Cl2.
The 1H and 13C NMR signals were assigned using 2-D (COSY, HETCOR, NOESY and HMBC) spectra shown in the Supplementary Materials. 2-D spectra were recorded using standard pulse sequences from Bruker pulse-sequence libraries.
In the mid-infrared region, the FT-IR spectra of MON-UR1-Na were recorded in CH2Cl2 solution, nujol/fluorolube mulls. A cell with silicon windows and wedge-shaped layers was used to avoid interferences (mean layer thickness, 170 μm). The spectra were taken with an IFS 113v FT-IR spectrophotometer (Bruker) equipped with a DTGS detector; resolution 2 cm–1, NSS=64. The Happ–Genzel apodization function was used.
Semi-empirical calculations
The PM5 quantum calculations were performed using the Win Mopac 2003 program at the semi-empirical level (Cache Work System Pro Version 5.04, Fujitsu, Beaverton, OR, USA). For MON-UR1-Na, the initial optimization of the structures was carried out using the molecular mechanics, extensive global minimum energy conformation search, with the Conflex/MM3 from WinMopac 2003 program. For the calculated, energetically the most favorable, structures of conformers corresponding with the global minimum points, further calculation of heat of formation was carried out using the PM5 quantum semi-empirical method.39
Microbiological analysis
Microorganisms used in this study were as follows: Gram-positive cocci: Staphylococcus aureus NCTC 4163, S. aureus ATCC 25923, S. aureus ATCC 6538, S. aureus ATCC 29213, Staphylococcus epidermidis ATCC 12228, Bacillus subtilis ATCC 6633, Bacillus cereus ATCC 11778, Enterococcus hirae ATCC 10541, Micrococcus luteus ATCC 9341, M. luteus ATCC 10240; Gram-negative rods: Escherichia coli ATCC 10538, E. coli ATCC 25922, E. coli NCTC 8196, Proteus vulgaris NCTC 4635, Pseudomonas aeruginosa ATCC 15442, P. aeruginosa NCTC 6749, P. aeruginosa ATCC 27863, Bordetella bronchiseptica ATCC 4617 and yeasts: Candida albicans ATCC 10231, C. albicans ATCC 90028, Candida parapsilosis ATCC 22019.
Hospital strains of S. aureus were isolated from different biological materials of patients of the Warsaw Medical University Hospital. Ten of these strains were methicillin-resistant S. aureus and ten other strains were methicillin-resistant S. epidermidis(. Owing to this resistance, these strains were tested for their sensitivity to the MONA and MON-UR1-Na. The other microorganisms used here were provided by the Department of Pharmaceutical Microbiology, Medical University of Warsaw, Poland.
Antimicrobial activity was examined by the disk-diffusion method under standard conditions using Mueller-Hinton II agar medium (Becton Dickinson, Heidelberg, Germany) for bacteria and RPMI agar with 2% glucose (Sigma), according to the Clinical and Laboratory Standards Institute (previously NCCLS) guidelines.40
Sterile filter paper disks (9 mm diameter, Whatman no. 3 chromatography paper) were dripped with tested compound solutions (in MeOH or MeOH/DMSO 1:1) to load 400 μg of a given compound per disk. Dry disks were placed on the surface of appropriate agar medium. The results (diameter of the growth inhibition zone) were read after 18 h of incubation at 35 °C. Compounds with recognized activity in disk-diffusion tests were examined by the agar dilution method to determine their MIC (Clinical and Laboratory Standards Institute).41 Concentrations of the agents tested in solid medium ranged from 3.125 to 400 μg ml−1. The final inoculum of all studied organisms was 104 CFU ml−1, except the final inoculum for E. hirae ATCC 10541, which was 105 CFU ml−1. The MIC values were read after 18 h of incubation at 35 °C. The data regarding the antimicrobial activity of the compounds are summarized in Tables 6 and 7.

The structures of MONA, MON-Na and MON-UR1-Na.
References
Ray, S. & Chaturvedi, D. Application of organic carbamates in drug design. Part 1: anticancer agents—recent reports. Drugs Fut. 29, 343–357 (2004).
Ray, S., Pathak, S. R. & Chaturvedi, D. Organic carbamates in drug development. Part II: antimicrobial agents—recent reports. Drugs Fut. 30, 161–180 (2005).
Westley, J. W. et al. Isolation of novel antibiotics X-14667A and X-14667B from Streptomyces cinnamonensis subsp. urethanofaciens and their characterization as 2-phenethylurethanes of monensins B and A. J. Antibiot. 34, 1248–1254 (1981).
Westley, J. W., Liu, C. M. & Evans, R. H. Preparation, properties and biological activity of natural and semisynthetic urethanes of monensin. J. Antibiot. 36, 1195–2000 (1983).
Rabaste, F., Jeminet, G., Dauphin, G. & Delort, A.- M. Na+ and K+ transport by 4-chlorophenylurethane-monensin in Enterococcus hirae de-energized and energized cells studied by 23Na-NMR and K+ atomic absorption. Biochim. Biophys. Acta. 1179, 166–169 (1993).
Rochdi, M. et al. Ionophore properties of monensin derivatives studied on human erythrocytes by 23Na NMR and K+ and H+ potentiometry: relationship with antimicrobial and antimalarial activities. J. Med. Chem. 39, 588–595 (1996).
Tanaka, R., Nagatsu, A., Mizukami, H., Ogihara, Y. & Sakakibara, J. Studies on chemical modification of monensin IX. Synthesis of 26-substituted monensins and their Na+ ion transport activity. Chem. Pharm. Bull. 49, 711–715 (2001).
Schildknechta, E. G., Siegela, D. & Richle, R. W. Antiparasitic activity of natural and semisynthetic monensin urethanes. Chemotherapy 29, 145–152 (1983).
Liu, C. M. & Westley, J. Monensin urethane derivatives. United States Patent 4263427 (http://www.freepatentsonline.com/4263427.pdf) (1981).
Riddell, F. G. Structure, conformation, and mechanism in the membrane transport of alkali metal ions by ionophoric antibiotics. Chirality 14, 121–125 (2002).
Duax, W. L., Smith, G. D. & Strong, P. D. Complexation of metal ions by monensin. Crystal and molecular structure of hydrated and anhydrous crystal forms of sodium monensin. J. Am. Chem. Soc. 102, 6725–6729 (1980).
Ward, D. L., Wei, K. T., Hoogerheide, J. C. & Popov, A. I. The crystal and molecular structure of the sodium bromide complex of monensin, C36H62O11·Na+Br−. Acta. Crystallogr. B 34, 110–115 (1978).
Pinkerton, M. & Steinrauf, L. K. Molecular structure of monovalent metal cation complexes of monensin. J. Mol. Biol. 49, 533–546 (1970).
Pangborn, W., Duax, W. & Langs, D. The hydrated potassium complex of the ionophore monensin A. J. Am. Chem. Soc. 109, 2163–2165 (1987).
Huczyński, A., Ratajczak-Sitarz, M., Katrusiak, A. & Brzezinski, B. Molecular structure of the 1:1 inclusion complex of monensin A sodium salt with acetonitrile. J. Mol. Struct. 832, 84–89 (2007).
Huczyński, A., Ratajczak-Sitarz, M., Katrusiak, A. & Brzezinski, B. Molecular structure of the 1:1 inclusion complex of monensin A lithium salt with acetonitrile. J. Mol. Struct. 871, 92–97 (2007).
Huczyński, A., Ratajczak-Sitarz, M., Katrusiak, A. & Brzezinski, B. Molecular structure of rubidium six-coordinated dihydrate complex with monensin A. J. Mol. Struct. 888, 224–229 (2008).
Martinek, T., Riddell, F. G., Wilson, C. & Weller, C. T. The conformations of monensin-A metal complexes in solution determined by NMR spectroscopy. J. Chem. Soc. Perkin Trans. 2, 35–41 (2000).
Westley, J. W. in Polyether Antibiotics. Naturally Occurring Acid Ionophores. Vol. 1, 1–20 (Marcel Dekker Inc., New York, 1982).
Westley, J. W. in Polyether Antibiotics. Naturally Occurring Acid Ionophores. Vol. 2, 51–86 (Marcel Dekker Inc., New York, 1983).
Pressman, B. C. In Antibiotics and Their Complexes 1–18 (Marcel Dekker Inc., New York, 1985).
Mollenhauer, H. H., Morre, D. J. & Rowes, R. D. Alteration of intracellular traffic by monensin; mechanism, specificity and relationship to toxicity. Biochim. Biophys. Acta. 1031, 225–246 (1990).
Awad, S. S. et al. Implementation of a methicillin-resistant Staphylococcus aureus (MRSA) prevention bundle results in decreased MRSA surgical site infections. Am. J. Surg. 198, 607–610 (2009).
Reygaert, W. Methicillin-resistant Staphylococcus aureus (MRSA): prevalence and epidemiology. Clin. Lab. Sci. 22, 111–114 (2009).
Bartlett, J. G. Methicillin-resistant Staphylococcus aureus infections. Top HIV Med. 16, 151–155 (2008).
Socrates, G. In Infrared and Raman Characteristic Group Frequencies: Tables and Charts 154, (John Wiley and Sons Ltd., UK, 2004).
Cutmore, E. A. & Hallam, H. E. Molecular configuration and interactions of the amide group: solvent effects on vibrational frequencies. Spectrochim Acta. 25A, 1767–1784 (1969).
Abramson, M. A. & Sexton, D. J. Nosocomial methicillin-resistant and methicillin-susceptible Staphylococcus aureus primary bacteremia: at what costs? Infect. Control Hosp. Epidemiol. 20, 408–411 (1999).
Mulvey, M. R. & Simor, A. E. Antimicrobial resistance in hospitals: how concerned should we be? Can. Med. Assoc. J. 180, 408–415 (2009).
Huczyński, A., Stefańska, J., Przybylski, P., Brzezinski, B. & Bartl, F. Synthesis and antimicrobial properties of Monensin A esters. Bioorg. Med. Chem. Lett. 18, 2585–2589 (2008).
Łowicki, D., Huczyński, A., Stefańska, J. & Brzezinski, B. Structural characterization and antibacterial activity against clinical isolates of Staphylococcus of N-phenylamide of monensin A and its 1:1 complexes with monovalent cations. Europ. J. Med. Chem. 45, 4050–4057 (2010).
Huczyński, A. et al. Synthesis of new semi-synthetic dipodands and tripodands from naturally occurring polyether ionophores. Tetrahedron Lett. 49, 5572–5575 (2008).
Łowicki, D. et al. Structural and antimicrobial studies of a new N-phenylamide of monensin A complex with sodium chloride. J. Mol. Struct. 923, 53–59 (2009).
Łowicki, D., Huczyński, A., Stefańska, J. & Brzezinski, B. Syntheses, structural and antimicrobial studies of a new N-allylamide of monensin A and its complexes with monovalent metal cations. Tetrahedron 65, 7730–7740 (2009).
Pedersen, D. S. & Rosenbohm, C. Dry column vacuum chromatography. Synthesis 16, 2431–2434 (2001).
Shusterman, A. J., McDougal, P. G. & Glasfeld, A. Dry-column flash chromatography. J. Chem. Educ. 74, 1222–1225 (1997).
Sheldrick, G. SHELXS-97, Program for Crystal Structure Solution (University of Göttingen, 1997).
Sheldrick, G. SHELXL-97, Program for Crystal Structure Refinement (University of Göttingen, 1997).
Przybylski, P., Huczyński, A. & Brzezinski, B. The structures of monensin A derivatives and its complexes with some monovalent cations studied by the AM1d, PM3 as well as PM5 semiempirical methods. J. Mol. Struct. 826, 156–164 (2007).
Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disc Susceptibility Tests; (Approved Standard M2-A9, Clinical and Laboratory Standards Institute, Wayne, PA, USA, 2006).
Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; (Approved Standard M7-A7, Clinical and Laboratory Standards Institute, Wayne, PA. USA, 2006).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Huczyński, A., Ratajczak-Sitarz, M., Stefańska, J. et al. Reinvestigation of the structure of monensin A phenylurethane sodium salt based on X-ray crystallographic and spectroscopic studies, and its activity against hospital strains of methicillin-resistant S. epidermidis and S. aureus. J Antibiot 64, 249–256 (2011). https://doi.org/10.1038/ja.2010.167
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2010.167
Keywords
This article is cited by
-
Finding disease modules for cancer and COVID-19 in gene co-expression networks with the Core&Peel method
Scientific Reports (2020)