Abstract
The bacterial genus Streptomyces is endowed with a remarkable secondary metabolism that generates an enormous number of bioactive small molecules. Many of these genetically encoded small molecules are used as antibiotics, anticancer agents and as other clinically relevant therapeutics. The rise of resistant pathogens has led to calls for renewed efforts to identify antimicrobial activities, including expanded screening of streptomycetes. Indeed, it is known that most strains encode >20 secondary metabolites and that many, perhaps most of these, have not been considered for their possible therapeutic use. One roadblock is that many strains do not express their secondary metabolic gene clusters efficiently under laboratory conditions. As one approach to this problem, we have used alleles of a pleiotropic regulator of secondary metabolism from Streptomyces coelicolor to activate secondary biosynthetic gene clusters in heterologous streptomycetes. In one case, we demonstrate the activation of pulvomycin production in S. flavopersicus, a metabolite not previously attributed to this species. We find that the absA1-engineered strains produced sufficient material for purification and characterization. As a result, we identified new, broad-spectrum antimicrobial activities for pulvomycin, including a potent antimicrobial activity against highly antibiotic-resistant Gram-negative and Gram-positive pathogens.
Similar content being viewed by others
Introduction
The bacterial genus Streptomyces is characterized by an abundant secondary metabolism that gives rise to numerous bioactive small molecules. More than 107 streptomycetes have been screened for antimicrobial activity, an effort that resulted in many of the antibiotics in current use.1 Given that there may be as many as 1026 species worldwide, it is likely that this genus produces many more compounds that are as yet unknown. Furthermore, the available genome sequences show that the known streptomycetes have genes for a greater abundance and diversity of secondary metabolites than has been revealed through most screening initiatives;2, 3, 4 for unknown reasons, most of these molecules are not produced efficiently in the laboratory. One challenge to renewed antimicrobial discovery is finding ways to enhance the production of these molecules.
The biosynthetic pathways that generate secondary metabolites are encoded in multigene clusters. Many of these are influenced by pleiotropically functioning regulators such as the absA locus of Streptomyces coelicolor,5 which encodes the sensor kinase AbsA1 and response regulator AbsA2 (Figure 1a). AbsA1 is both an AbsA2 phosphotransferase and an AbsA2∼P phosphatase.6, 7 AbsA2∼P represses several biosynthetic gene clusters and absA1 mutations that impair AbsA2 phosphorylation enhance the expression of these clusters.6, 7 absA-like operons are found primarily in actinomycetes and are often embedded in the biosynthetic gene clusters of secondary metabolites,8 perhaps suggesting that the regulation of secondary metabolism by this subfamily is a common phenomenon. We previously described two alleles of absA1 that enhance antibiotic production in S. coelicolor, the point mutant absA1H202A and the in-frame deletion mutant absA1del3/4.7, 8
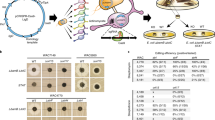
Rationale for remodeling secondary metabolism with alleles of absA1. (a) In S. coelicolor, AbsA1 phosphorylates AbsA2 and dephosphorylates AbsA2∼P. AbsA2∼P binds and represses some secondary metabolic gene clusters. (b) To remodel secondary metabolism, we expressed S. coelicolor absA1 genes in heterologous streptomycetes, assuming that this might counteract the effects of endogenous AbsA1 proteins (AbsA1e) dephosphorylating the endogenous AbsA2 protein (AbsA2e) and enhancing the expression of some secondary metabolic genes. Examples of this effect are shown in panels c–f. EV refers to a strain bearing the empty vector throughout. The wild isolate Wat#1 exhibited enhanced antimicrobial activity against B. subtilis (c), as well as the accumulation of a blue pigment (d) when it expressed absA1del3/4. (e) A culture supernatant of S. griseochromogenes bearing the absA1H202A allele exhibited enhanced antibiotic activity as demonstrated by a zone of inhibited growth in a lawn of Micrococcus luteus. (f) A strain of S. flavopersicus bearing the absA1H202A allele produced a novel antimicrobial activity against B. cenocepacia.
In this study, we have used alleles of S. coelicolor absA1 to alter secondary metabolism in heterologous streptomycetes. We screened the engineered strains for activities that inhibit the growth of antibiotic-sensitive laboratory strains of Escherichia coli, Bacillus subtilis, Micrococcus luteus and Staphylococcus aureus, finding that absA1 could activate new antimicrobial activity in roughly half of the streptomycetes. To prioritize these activities for a more fulsome characterization, we also screened against the highly antibiotic-resistant opportunistic pathogens Burkholderia cenocepacia and Pseudomonas aeruginosa, reasoning that molecules that inhibit their growth would be of greater interest. In this way, we identified one absA1-induced molecule having significant antimicrobial activity against antibiotic-resistant Gram-positive and Gram-negative bacteria. We will argue that this approach could be used to activate secondary metabolism in previously isolated Streptomyces species and in new soil isolates, and that this could accelerate the discovery of new lead compounds having therapeutic use.
Materials and methods
Genetic engineering and growth of bacteria for antimicrobial screening
Molecular genetic manipulation of streptomycetes was carried out using standard procedures.9 Plasmids pSET152, pabsA1, pabsA1H202A and pabsA1del3/4 were introduced into the streptomycetes listed in Table 1 by conjugation from ET12567/pUZ8002. Apramycin-resistant exconjugants were purified by growth in the presence of 50 μg ml−1 apramycin and spore stocks were prepared. To prepare culture supernatants, each strain was streaked on Bennett's agar media supplemented with 50 μg ml−1 apramycin for 6 days at 30 °C, followed by inoculation of spores from a single colony into 3 ml Bennett's broth in culture tubes with glass beads for dispersion of mycelia and incubated (30 °C) for 5 days. Cultures were then centrifuged at 10 000 r.p.m. for 10 min and 100 μl supernatant was used for LC-MS studies. For antimicrobial screening, spots of each strain, containing equivalent numbers of spores, were grown on Bennett's agar and Oxoid nutrient agar (ONA) supplemented with 12 mM calcium nitrate. After 2 days of growth, the plates were overlaid with 3 ml of soft agar (1:1 Bennett's or ONA agar/broth), containing a 1/100 dilution of an overnight culture of the bioassay indicator strain. Plates were then incubated for 16–27 h at 27 °C and antimicrobial activity was visualized as a zone of clearing in the soft agar.
Biochemical characterization of secondary metabolites
Spores were thoroughly streaked on 10 cm plates containing Bennett's agar supplemented with 50 μg ml−1 apramycin for 6 days at 30 °C in the dark. Agar was chopped to fine pieces and secondary metabolites were extracted using 25 ml acetone and 12 h of shaking at 4 °C. For 15 min, the supernatant was concentrated by centrifugation at 8000 r.p.m. to a final volume of 1 ml. Concentrated acetone extracts (50 μl) were analyzed by LC/ESI-MS by using an Agilent 1100 Series LC system (Agilent Technologies Canada, Mississauga, ON, Canada) and a QTRAP LC/MS/MS system (Applied Biosystems/MDS Sciex, Streetsville, ON, Canada). Analytical reverse-phase HPLC was performed using a C18 column (3 μm, 120 A, 4.6 × 10 mm; Dionex Canada Ltd., Oakville, ON, Canada) at a flow rate of 1 ml min−1, using isocratic 5% solvent B (0.05% formic acid in acetonitrile) and 95% solvent A (0.05% formic acid in water), for 5 min, followed by a linear gradient to 97% B over 20 min. ESI-MS data were obtained by screening in positive and negative ion mode. MS/MS data were acquired using Information Dependent Acquisition in the mass range of 50–1700 Da and the following settings: DP-80, EP-10, rolling collusion energy, CUR 12.0, CAD set at medium, IS-4500, TEM 100 °C, CSI 30 and interface heater off.
Purification and novel antimicrobial activities of pulvomycin
For analytical-scale analysis of anti-Burkholderial activity, agar plugs of S. flavopersicus expressing absA1H202A were lyophilized overnight and subjected to a three-phase solvent extraction composed of n-hexane-methyl acetate-acetonitrile-water (4:4:3:4, v/v/v/v). The active fraction was split into two aliquots and subjected to TLC on two TLC silica plates (ALUGRAM SILG/UV254, 0.2 mm, Camlab Ltd, Cambridge, UK) with separation using a chloroform/methanol/water (65:25:4, v/v/v) mobile phase. The spots on one plate were revealed under long (350 nm) and short (254 nm) wavelength UV light, and outlined on one TLC plate. The second TLC plate was dried, and overlaid with 20 ml of soft agar (1:1 ONA/ON broth) containing an overnight culture of B. cenocepacia diluted 1/100 and incubated overnight at 27 °C. To develop the plate, 1.5 ml of 5 mg ml−1 Thiazolyl Blue Tetrazolium Bromide (MTT, Sigma, Oakville, ON, Canada) was added to the agar surface and incubated for 1 h at 37 °C. The spot on the plate that was active against B. cenocepacia was identified by a yellow zone in the presence of MTT. For preparative scale analysis, pulvomycin was isolated from 600 g of fresh S. flavopersicus mycelium expressing the H202A allele of absA1 cultured in Bennett's broth for 5 days. Extraction from mycelia was carried out using 800 ml of dichloromethane/methanol (3:1, v/v), with continuous stirring at 4 °C overnight in the dark. The solvent was filtered, concentrated by rotary evaporation at room temperature and lyophilized. The crude extract was dissolved in a small volume of acetone for further purification by TLC using a chloroform/methanol (9:1, v/v) solvent system. The final product was recovered from silica by dissolving it in acetone and shaking for 2 h at 4 °C, followed by centrifugation at 10 000 r.p.m. for 10 min. LC-MS was used to confirm the purity and integrity of the molecule. The compound was dried and redissolved in methanol and stored at −20 °C. The yield of pulvomycin was 12.8 mg, as determined using the molar absorbance coefficient (AM) of 74582 at 320 nm in methanol.
MIC determination
MIC values were determined following Clinical Laboratory Standards Institute guidelines.
Results
Engineering streptomycetes to produce new antimicrobial activities
If pleiotropic regulation of secondary metabolism by absA-like operons occurs in other streptomycetes, it should be possible to use wild-type or kinase-defective alleles of the S. coelicolor AbsA1 protein (AbsA1Sc; see Figure 1b) to alter the phosphorylation state of the endogenous AbsA2 protein (AbsA2e) and thereby enhance the expression of some or all biosynthetic genes. We therefore introduced the wild-type, H202A and del3/4 alleles of absA1Sc into 15 Streptomyces strains, along with the empty vector pSET152. The engineered strains included nine characterized producers of known secondary metabolites and six environmental soil isolates (Table 1).
Of the 15 empty vector-containing control strains, 13 inhibited the growth of one or more E. coli, B. subtilis, M. luteus and S. aureus, as revealed by a zone of inhibited growth (Supplementary Figure 1). The strains exhibited very limited effects on both P. aeruginosa and B. cenocepacia, consistent with the high level of antibiotic resistance associated with these bacteria.10, 11
The introduction of absA1 alleles altered the secondary metabolism of many of the streptomycetes, as demonstrated by increased antimicrobial activity (revealed by enlarged zones of inhibition against target bacteria) or, in at least one case, the activation of a blue pigment (Figures 1c–f, Table 1 and Supplementary Figure 1). In some cases, all three absA1 alleles had the same effect, although in others, the stimulatory effect was restricted to the kinase-defective alleles. We confirmed that the empty vector did not influence antimicrobial activities in a subset of the parent stains; therefore, the effects described above were absA1 dependent and not the result of genetic instability.12
Two strains, Va#1b and Va#39, did not produce any detectable antimicrobial activity against any of the target bacteria, regardless of growth conditions or the introduction of absA1 genes. Of the strains exhibiting absA1-induced antimicrobial activities, three were soil isolates (Cu#2B, Ja#2b and KC2#14) and nine were previously characterized producers of known antibiotics. One of the most exciting outcomes was the effect of absA1 on S. flavopersicus NRRL2820, wherein the gene conferred the ability to inhibit the growth of B. cenocepacia. Spectinomycin, the only previously reported S. flavopersicus antibiotic, had no antimicrobial activity against B. cenocepacia (data not shown).
Biochemical characterization of the induced secondary metabolites
To view secondary metabolite production directly, we compared the patterns of small molecule accumulation in the empty vector- and absA1Sc-containing strains using HPLC, mass spectrometry and UV absorbance analysis. Culture supernatants of S. griseochromogenes contained a small molecule having m/z=422.8, corresponding to the peptidyl nucleoside antibiotic blasticidin S, a known product of this species13 (Figures 2a and b). The yield of this molecule was enhanced ∼5-fold by absA1Sc and likely accounts for the absA1H202A-induced antimicrobial activity in this supernatant (Figure 1c).
Secondary metabolite production by streptomycetes strains containing the empty vector (red trace) and strains containing the absA1H202A allele (blue trace). (a) LC analysis of culture supernatant of a strain of S. griseochromogenes engineered with the absA1H202A gene produces one peak (2.4 min) that is greatly reduced in the empty vector-containing strain. MS analysis of this peak (b) shows a m/z+ of 422.8 Da, consistent with the antibiotic blasticidin S. (c) A strain of S. flavopersicus containing absA1H202A produces at least three extracellular metabolites (18.71, 20.48 and 21.99 min) at greatly elevated levels relative to the vector control. The largest of these peaks was purified and identified as the antibiotic pulvomycin.
As a result of its absA1H202A-stimulated antimicrobial activity against B. cenocepacia, we were particularly interested in the metabolites induced in S. flavopersicus NRRL2820. The absA1-containing strain produced at least three absA1-stimulated molecules that could be recovered in hydrophobic extracts of cell surfaces (Figure 2c). These molecules had m/z=212 (18.71 min, Amax=260 nm), 837 (20.48 min, Amax=266, 276 and 322 nm) and 397 (21.99 min, Amax=256 nm). There is no published report of molecules having these masses as S. flavopersicus natural products. We did not directly observe spectinomycin (mass=332 Da), a known S. flavopersicus NRRL2820 metabolite,14 by this strain, although we confirmed that the one known spectinomycin biosynthetic gene, spcR, was present in our strain in wild-type form (Yu and JR Nodwell, unpublished data).
To determine which of the S. flavopersicus molecules was responsible for the inhibition of Burkholderia growth, we purified the active compound from this mixture using a three-phase organic extraction and preparative TLC (Figure 3a). Chromatographic analysis revealed that it corresponded to the most abundant of the absA1-induced species (Figure 3b).
Purification and identification of the anti-Burkholderia compound. (a) Anti-Burkholderia activity was extracted from absA1H202A expressing S. flavopersicus NRRL2820 cells using a three-phase procedure (see Materials and methods section). Each phase (upper (UP), middle (MP) and lower (LP)) was applied to filter disks and was assessed for anti-Burkholderia activity; the activity was clearly restricted to the middle phase. (b) We subjected the middle phase to TLC, and overlaid the plate with soft agar containing Burkholderia. The spot containing the antibiotic was identified by a zone of B. cenocepacia clearing (indicated by arrow). (c) The upper panel shows the LC trace of the TLC-purified peak eluted at 20.58 min, corresponding to the absA1-stimulated metabolite with the strongest UV response (blue trace, 20.48 min, in Figure 2c). The lower panel shows the UV spectral analysis of the purified peak with UV max at 266, 274 and 322 nm, all characteristic of pulvomycin. (d) NMR analysis revealed that the structure of the purified compound is identical to the rare metabolite pulvomycin, purified previously from S. netropsis. A full colour version of this figure is available at The Journal of Antibiotics journal online.
We were unable to solve the structure of the purified molecule using mass spectrometry and therefore resorted to proton NMR and 13C NMR (see supplementary Figures 2–9 and Supplementary Tables 1–3). In this way, we demonstrated that the activity was identical to pulvomycin15 (Figure 3c) isolated previously from S. netropsis. Pulvomycin is an inhibitor of the translation elongation factor EF-Tu.16 The purified molecule exhibited physical properties identical to those of pulvomycin (m/z=837, Amax=266, 276 and 322 nm) and is clearly evident as an absA1-stimulated peak (Figure 2c).
We therefore used an absA1H202A-expressing S. flavopersicus strain to purify sufficient quantities for functional analysis and found that the antimicrobial activity (Table 2) was superior to that reported previously.17 Consistent with our screening results, we found that the MIC against B. cenocepacia and B. vietnamiensis was 8 μg ml−1. The purified molecule also exerted activity (MICs of 16–32 μg ml−1) against P. aeruginosa and Acinetobacter baumannii (ATCC 17978), two other opportunistic pathogens that are highly antibiotic resistant. Its activity against Gram-positive pathogens was even more impressive: MICs against clinical isolates of methicillin-resistant S. aureus and vancomycin-resistant Enterococcus (vanB) were 2–4 μg ml−1. These antimicrobial activities have not been reported for pulvomycin previously.
Discussion
These data demonstrate that the S. coelicolor absA1 gene can significantly alter secondary metabolism when expressed in many heterologous streptomycetes, activating the production of secondary metabolites that are otherwise expressed poorly or not at all. The most logical explanation for this is that the strains that responded have endogenous absA operons that pleiotropically regulate secondary metabolism. According to this scenario, the expression of the S. coelicolor absA1 gene caused reduced levels of phosphorylation of the endogenous AbsA2 protein, relieving repression of its target promoters (Figure 1).
Indeed, a search of the sequenced actinomycetes genomes reveals sensor kinases having high-sequence identity to AbsA1 (P-values between e−20 and e−45) in all of them. Using the most stringent criteria, including both high-sequence identity and the atypical arrangement of transmembrane domains that we observed for AbsA1,8 we found two putative AbsA1 orthologs in S. roseosporus (SSIG_02156.3 and SSIG_03787.3) and one each in S. scabies (SCAB18851), S. kasugaensis (KasO), Streptomyces species Mg1 (SSAG_02972.2), S. griseus (SGR3297), the actinomycete Saccharopolyspora erythraea (SACE_7062) and in the currently unpublished S. venezuelae genome sequence (Mervyn Bibb, personal communication). The absA1-like genes were all proximal to absA2-like genes. The S. coelicolor AbsA1 protein could have either directly dephosphorylated the endogenous AbsA2∼P protein, assembled into inactive heterodimers with the endogenous AbsA1 protein, or could have disrupted the (currently unknown) signaling pathway that controls the balance of the AbsA1 kinase and phosphatase activities.
We were intrigued by the observation that absA1 alleles caused reduced levels of some antimicrobial activities and biochemical peaks Figure 2a. This was unexpected on the basis of the existing model of AbsA1 function in S. coelicolor, which suggests that AbsA2∼P represses pathway-specific activators. It is known, however, that streptomycetes can assemble similar sets of regulators into different networks.18 In some strains therefore, absA-like genes may repress some genes and activate others. Alternatively, perhaps some secondary metabolic gene clusters are repressed by proteins that are in turn repressed by AbsA2e∼P.
However, the absA1 gene exerted these effects; it is clear that this approach could have significant use in the discovery of antibiotics and other useful molecules. In this relatively modest screen, we have induced new antimicrobial activity in at least 10 streptomycetes and identified a previously unknown activity for pulvomycin. Indeed, pulvomycin is not commercially available and its remarkable antimicrobial activity against highly resistant pathogens was unknown until this study. In addition to discovering new antibiotics, our approach could facilitate the reinvestigation of molecules that have been underappreciated and discarded because they were difficult to purify from wild-type strains.
There are many possible applications of the study we have described here. It is clear that the expression of wild-type or mutated pleiotropic regulators can be used to generate collections of streptomycetes in which cryptic biosynthetic gene clusters have been activated. Indeed, there are other pleiotropic regulators of secondary metabolism in S. coelicolor19, 20 and it is likely that some of them could be adapted to this purpose as well. Streptomycetes engineered in this way could be screened directly for new antimicrobial or other activities (as in Table 1). Alternatively, those induced metabolites exhibiting the greatest biochemical novelty (see Figure 2 and Table 2) could simply be purified to improve the chemical diversity of natural product libraries. We suggest as well that screening for antimicrobial activities requires the inclusion of highly resistant target bacteria such as B. cenocepacia and P. aeruginosa. In our case, although screening with laboratory strains revealed the largest number of absA1-activated antimicrobial activities, the inclusion of B. cenocepacia narrowed our interest to the one activity of greatest interest. This in turn revealed that the previously identified molecule pulvomycin inhibits the growth of several highly resistant pathogenic bacteria. It is our hope that screening engineered streptomycetes against both antibiotic-sensitive and -resistant target strains, along with innovations from other laboratories,21 will help to fuel a much needed renaissance in antibacterial discovery.
References
Baltz, R. H. Antimicrobials from Actinomycetes: Back to the Future. Microbe. 2, 6–7 (2007).
Bentley, S. D. et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147 (2002).
Ikeda, H. et al. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21, 526–531 (2003).
Ohnishi, Y. et al. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 190, 4050–4060 (2008).
Adamidis, T., Riggle, P. & Champness, W. Mutations in a new Streptomyces coelicolor locus which globally block antibiotic biosynthesis but not sporulation. J. Bacteriol. 172, 2962–2969 (1990).
Brian, P., Riggle, P. J., Santos, R. A. & Champness, W. C. Global negative regulation of Streptomyces coelicolor antibiotic synthesis mediated by an absA-encoded putative signal transduction system. J. Bacteriol. 178, 3221–3231 (1996).
Sheeler, N. L., MacMillan, S. V. & Nodwell, J. R. Biochemical activities of the absA two-component system of Streptomyces coelicolor. J. Bacteriol. 187, 687–696 (2005).
McKenzie, N. L. & Nodwell, J. R. Transmembrane topology of the AbsA1 sensor kinase of Streptomyces coelicolor. Microbiology 155, 1812–1818 (2009).
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F. & Hopwood, D. A. Practical Streptomyces genetics (The John Innes Foundation, Norwich, UK, 2000).
Guglierame, P. et al. Efflux pump genes of the resistance-nodulation-division family in Burkholderia cenocepacia genome. BMC Microbiol. 6, 66 (2006).
Poole, K. & Srikumar, R. Multidrug efflux in Pseudomonas aeruginosa: components, mechanisms and clinical significance. Curr. Top Med. Chem. 1, 59–71 (2001).
Volff, J. N. & Altenbuchner, J. Genetic instability of the Streptomyces chromosome. Mol. Microbiol. 27, 239–246 (1998).
Cone, M. C., Petrich, A. K., Gould, S. J. & Zabriskie, T. M. Cloning and heterologous expression of blasticidin S biosynthetic genes from Streptomyces griseochromogenes. J. Antibiot (Tokyo) 51, 570–578 (1998).
Mason, D. J., Dietz, A. & Smith, R. M. Actinospectacin, a new antibiotic. I. Discovery and biological properties. Antibiot. Chemother. 11, 118–122 (1961).
Smith, R. Structure revision of the antibiotic pulvomycin. J Am. Chem. Soc. 107, 2849–2859 (1985).
Wolf, H., Assmann, D. & Fischer, E. Pulvomycin, an inhibitor of protein biosynthesis preventing ternary complex formation between elongation factor Tu, GTP, and aminoacyl-tRNA. Proc. Natl Acad. Sci. USA 75, 5324–5328 (1978).
Selva, E. et al. Antibiotic GE2270 a: a novel inhibitor of bacterial protein synthesis. I. Isolation and characterization. J. Antibiot. (Tokyo) 44, 693–701 (1991).
Chater, K. F. & Horinouchi, S. Signalling early developmental events in two highly diverged Streptomyces species. Mol. Microbiol. 48, 9–15 (2003).
van Wezel, G. P., McKenzie, N. L. & Nodwell, J. R. Chapter 5. Applying the genetics of secondary metabolism in model actinomycetes to the discovery of new antibiotics. Methods Enzymol. 458, 117–141 (2009).
Rigali, S. et al. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 9, 670–675 (2008).
Hosaka, T. et al. Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat. Biotechnol. 27, 462–464 (2009).
Acknowledgements
We thank Susan Jensen and Marie Elliot for helpful comments on this paper. We also thank Mervyn Bibb for communicating unpublished information with us. Work in JRN's laboratory was supported by the Natural Science and Engineering Research Council (165846) and by JNE Biotech Inc. (Hamilton, Ontario). NLM was supported by a Canadian Institutes of Health Research Canada Graduate Scholarship (CGD—80406). Work in GDW's laboratory was supported by the Canadian Institutes for Health Research (MT-14981) and the Natural Science and Engineering Research Council.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
McKenzie, N., Thaker, M., Koteva, K. et al. Induction of antimicrobial activities in heterologous streptomycetes using alleles of the Streptomyces coelicolor gene absA1. J Antibiot 63, 177–182 (2010). https://doi.org/10.1038/ja.2010.13
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2010.13
Keywords
This article is cited by
-
Identification of pulvomycin as an inhibitor of the futalosine pathway
The Journal of Antibiotics (2021)
-
Production of pikromycin using branched chain amino acid catabolism in Streptomyces venezuelae ATCC 15439
Journal of Industrial Microbiology and Biotechnology (2018)
-
Identification of butenolide regulatory system controlling secondary metabolism in Streptomyces albus J1074
Scientific Reports (2017)
-
Competition and co-regulation of spirotoamide and tautomycetin biosynthesis in Streptomyces griseochromogenes, and isolation and structural elucidation of spirotoamide C and D
The Journal of Antibiotics (2017)
-
Metabolic perturbation to enhance polyketide and nonribosomal peptide antibiotic production using triclosan and ribosome-targeting drugs
Applied Microbiology and Biotechnology (2017)