Abstract
Escherichia coli spans a genetic continuum from enteric strains to several phylogenetically distinct, atypical lineages that are rare in humans, but more common in extra-intestinal environments. To investigate the link between gene regulation, phylogeny and diversification in this species, we analyzed global gene expression profiles of four strains representing distinct evolutionary lineages, including a well-studied laboratory strain, a typical commensal (enteric) strain and two environmental strains. RNA-Seq was employed to compare the whole transcriptomes of strains grown under batch, chemostat and starvation conditions. Highly differentially expressed genes showed a significantly lower nucleotide sequence identity compared with other genes, indicating that gene regulation and coding sequence conservation are directly connected. Overall, distances between the strains based on gene expression profiles were largely dependent on the culture condition and did not reflect phylogenetic relatedness. Expression differences of commonly shared genes (all four strains) and E. coli core genes were consistently smaller between strains characterized by more similar primary habitats. For instance, environmental strains exhibited increased expression of stress defense genes under carbon-limited growth and entered a more pronounced survival-like phenotype during starvation compared with other strains, which stayed more alert for substrate scavenging and catabolism during no-growth conditions. Since those environmental strains show similar genetic distance to each other and to the other two strains, these findings cannot be simply attributed to genetic relatedness but suggest physiological adaptations. Our study provides new insights into ecologically relevant gene-expression and underscores the role of (differential) gene regulation for the diversification of the model bacterial species.
Similar content being viewed by others
Introduction
The rapidly increasing availability of bacterial genomes is providing detailed insights into bacterial diversity and their evolutionary history, which is increasingly questioning traditional taxonomic categorizations. New approaches based on whole genome comparisons such as Average Nucleotide Identity (ANI) have emerged and combined with eco-physiological concepts (for example, ecotype concept (Van Valen, 1976)) could lead to a more holistic bacterial species definition (Rosselló-Mora and Amann, 2001; Konstantinidis et al., 2006; Cohan and Kopac, 2011; Caro-Quintero and Konstantinidis, 2012). Escherichia coli exemplifies the complexities of the current species definition issue. It spans a genetic continuum from the human associated enteric E. coli to several atypical clades that are frequently detected in the environment and in birds (Clermont et al., 2011). All clades share the same core genes (that is, atypical strains exhibit all genes considered typical for enteric E. coli) and show remarkable biochemical similarities, which left them categorized as the same species (Walk et al., 2009). Subsequently, a taxonomic placement of these atypical clades as different species was proposed because (i) phylogenetic analysis cluster human enteric strains together and apart from atypical strains, (ii) several genes that are considered important for adaptation to distinct primary habitats (the human gut versus the environment) are specifically enriched in the individual groups and (iii) the exchange of genetic material has been shown to occur predominantly within strains sharing the same primary habitat (Luo et al., 2011).
The relative importance of regulatory versus coding-sequence evolution in adaptation and diversification has long been debated, and much recent work has attempted to answer this question using various organisms (King and Wilson, 1975; Stern and Orgogozo, 2008). By now it is well-recognized that cis-regulatory change is a key process in evolution for the origin of distinct phenotypes in eukaryotes (Enard et al., 2002). In bacteria, the importance of gene-expression as a governing factor for evolution, habitat adaptation and diversification is recognized as well (c.f. Philippe et al., 2007) and initial data from a pioneer study that correlated global protein expression profiles with evolutionary relatedness in the Shewanella genus are available (Konstantinidis et al., 2009). However, the field is largely unexplored and global, omics-based data specifically investigating the link between gene regulation and phylogeny under different culture conditions are needed to understand the role of gene expression in bacterial diversification.
The aim of the present study was to investigate the association between gene regulation and phylogeny in E. coli, and to specifically explore whether gene expression contributes to diversification in this taxon. To this end, we compared global gene expression profiles of four strains of E. coli; the laboratory strain MG1655, a model commensal (IAI1) and two atypical strains, which were recovered from extra-intestinal habitats (environmental strains) and represent distinct lineages (clade IV and clade V) (Luo et al., 2011). All strains show similar genetic relatedness with an ANI of 92–93%, except for the laboratory and commensal strains that are closer related (98.6% ANI; Supplementary Table S1). The strains were grown under three different conditions namely unlimited growth (exponential batch growth), carbon-limited growth (chemostat) and starvation (no nutrient flow to chemostat culture for four hours), and their global gene expression profiles were compared to explore whether there are trends toward adaptation to the gut, laboratory and extra-intestinal environments.
Materials and methods
Bacterial strains and growth conditions
The E. coli laboratory strain K12 (MG1655), a commensal model strain (IAI1) and two distantly related atypical E. coli strains (TW11588—Clade IV and TW09308—Clade V) isolated from the environment namely soil and freshwater beach, respectively (Luo et al., 2011)), were used in the study. Phylogenetic relationships among the strains were measured as the ANI of all pairwise comparisons as described previously (Goris et al., 2007) (Supplementary Table S1). A minimal growth medium as described in the study by Ihssen and Egli (2004) was used for all experiments. Bacterial stock cultures were streaked onto agar plates and incubated overnight. One colony was then transferred into 20 ml minimal medium, grown at 37 °C (250 r.p.m.) over night culture (ONC) and served as the inoculum for experiments. For continuous culture experiments we designed and constructed 500 ml bioreactors according to the study by Huwiler et al., (2012) that were half-filled with medium (0.5 g glucose per l) and incubated at 37 °C in a temperature controlled water bath. Before continuous cultivation (dilution rate=0.25), 1–2 ml of the ONC was transferred and grown in batch-mode until reactors became visibly turbid. Subsequently, cells were grown to steady-state (defined as constant optical density over time) and harvested for experimentation. For starvation experiments the medium flow was stopped during steady-state and bacteria were collected after 4 h. To avoid gene-expression signatures of stationary cells from the ONC, batch cultures (1000 ml Erlenmeyer flasks containing 100 ml of pre-warmed medium (1 g glucose per l); 37 °C; 250 r.p.m.)) were inoculated with 5 ml of an exponentially growing pre-culture that derived from the ONC.
RNA extraction and sequencing
Harvested cells (4 × 6 ml) were immediately combined with 6 ml RNAlater (Life Technologies, Grand Island, NY, USA), centrifuged for 15 min at 12 000 r.p.m., washed with 1 ml RNAlater (3 min at 15 000 r.p.m.), re-suspended in 0.5 ml RNAlater and stored at −20 °C. For RNA extraction the RiboPure-Bacteria Kit (Life Technologies) was used according to the manufacturer’s instructions. To achieve high RNA yields several reactions for each sample were done in parallel and pooled at the end of the procedure. An additional DNase treatment step with TURBO DNase (Life Technologies) was included to assure no genomic DNA contamination. Messenger RNA was enriched using the RiboMinus Bacteria Kit (Life Technologies) according to the manufacturer’s instructions. RNA quality was confirmed with the Bioanalyzer system (Agilent Technologies, Santa Clara, CA, USA) and sequencing (50 cycles, pooling eight bar-coded samples per lane) was performed on the Illumina HiSeq platform (Illumina, San Diego, CA, USA) at the Research Technology Support Facility (RTSF) at Michigan State University.
Data pre-processing
Sequence reads passing quality filtering were trimmed to remove the 3′ end low quality segments flagged by Read Segment Quality Control Indicator. Reads from each sample were mapped to their respective source genomes using Bowtie (Langmead et al., 2009) with alignment parameters (-n 2-e 70-l 28 –best). Reads that mapped to unique genomic sites were tallied and summarized for each protein-coding gene feature annotated for the corresponding reference genome (Luo et al., 2011). A high percentage of reads mapped back to genomes (median: 98.4%), except for strain TW11588 where mapping percentages were considerably lower (median: 89.6%). Analysis revealed that the majority of these unmapped reads of TW11588 were of ribosomal RNA origin (median: 90.2%) and therefore, did not affect follow-up analyses where only the protein-coding genes were considered. Additional checks were performed by cross-validating gene features against RAST annotations of these genomes to remove additional non-protein-coding genes. Read count data from technical replicates were pooled for each biological sample.
Statistical analysis
Comparisons of genes between treatments for differential expression were performed using the DESeq package (Anders and Huber 2010) in R, with the Negative Binomial Distribution for better modeling the read count variability over low to high dynamic range between biological replicates. For gene counts, the median-based method was used to normalize effective library sizes and a false discovery rate of 5% was used as the cutoff for calling differential expression. The normalized library counts computed by DESeq and the nucleotide base length of each gene feature were used to calculate Reads Per Kilobase of gene per Million mapped reads (RPKM). Each gene was categorized based on its gene ortholog among 25 genomes based on (Luo et al., 2011): core genes, present in all 25 strains minus one; commonly shared genes, present in all four strains; genes enriched in each group (enteric versus environmental; group-specific genes) based on (Luo et al., 2011); unique genes (unique to a strain), and all remaining genes (others). The nucleotide identity of each ortholog pair was computed as described in (Goris et al., 2007).
Dendrograms were built from RPKM data using the Vegan statistics package in R, where value of a gene in any genome was set to zero if this gene ortholog is not present in this genome. Data were first scale-transformed (Wisconsin) before dissimilarity calculations with the Euclidean formula for each sample pair and final hierarchical clustering (‘hclust’). The functional category(s) of each gene was determined by using RAST subsystem annotations (http://rast.nmpdr.org) of the uploaded genome sequences, supplemented by MG-RAST (http://metagenomics.anl.gov) for gene features, for which subsystem assignments were lacking from RAST. Genes linked to uptake/catabolism of carbon substrates and stress response are of specific interest for the culture conditions applied and detailed analysis focusing on those features was performed based on the study by Ihssen et al. (2007).
Results
Overall expression data
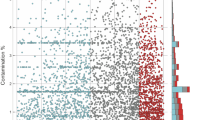
The basic growth parameters maximum specific growth rate (μmax) and OD yield were similar for all strains under all culture conditions (Supplementary Table S2). Expression levels of individual gene sets were highest for the core, followed by commonly shared (noncore) genes and other sets namely unique, others and group-specific genes (Supplementary Figure S1). Overall, we detected a positive correlation between ortholog frequency in 25 representative E. coli genomes (based on the study by Luo et al. (2011)) and expression level (Figure 1). In other words, the wider a gene was distributed, the higher was its expression level. Three distinct clusters were detected with lowest expression levels for genes present in <60% of E. coli strains, median values for genes with 60 to 90% orthologous frequency and a steep increase >90% (Figure 1).
Expression level correlated with ortholog frequency (the proxy of gene conservation level), that is, percentage of E. coli strains encoding that gene based on the study by Luo et al. (2011). Each dot refers to all genes of a specific orthologous frequency group where expression levels are based on the median of all maxima (maximum values (average of replicates) observed in any strain under any condition). Three distinct clusters are indicated, where the two uppermost points represent E. coli core genes.
Differential gene expression analysis demonstrated large physiological differences between states (culture conditions) where 42.8% (batch versus chemostat), 70.7% (batch versus starvation) and 66.9% (chemostat versus starvation) of all genes were differentially expressed (Supplementary Table S3). Replicate samples showed very high reproducibility with a median R2 of 0.99 (range 0.94–1; data not shown). Overall expression level shapes, that is, overall transcript distribution patterns, were very similar between states and strains (Supplementary Figures S2 and S3), demonstrating that observed differences were indeed due to specific bacterial responses for individual culture conditions. Typical known global responses to each culture condition were detected in all strains. For instance, compared with batch cultures, chemostat growth and starvation resulted in upregulation of numerous high-affinity uptake systems for diverse carbon substrates, together with catabolic enzymes for their degradation (Supplementary Figure S4). Upregulation was less pronounced in starvation, and direct comparisons between chemostat and starvation demonstrated a downregulation of many carbonaceous uptake/catabolism enzymes after cells entered starvation (Supplementary Figure S4). In addition, both carbon-limited growth and starvation triggered an increased expression of various stress defense mechanisms, which were particularly pronounced in the latter state (Supplementary Figure S5). Starvation was further characterized by gene expression patterns indicating dwarfing/autophagy (for example, upregulation of fatty acid metabolizing fad genes and several protease genes such as clp), together with a downregulation of central growth related genes including ribosomal proteins and ATPase subunits among others (Supplementary Data set S1). Efflux pumps and biofilm formation genes were specifically upregulated in starvation (Supplementary Data set S1).
Gene regulation and coding-sequence evolution are connected
Highly differentially expressed genes between all pairs (P<0.01; greater than and equal to twofold difference) showed a significantly lower nucleotide identity compared with other genes, demonstrating that the degree of sequence conservation is correlated with gene expression (Figure 2). This pattern was more pronounced in growth states (batch and chemostat) compared with starvation (results of all individual pairs are presented in Supplementary Figure S6). Interestingly, analysis of horizontally transferred genes between environmental clades (core genes only), based on the study by Luo et al., (2011), revealed no significant differences in both overall expression level and expression differences between strains compared with other core genes, suggesting that transfer was neutral with respect to gene regulation (data not shown).
Nucleotide identity of highly differentially expressed genes (greater than and equal to twofold; gray box plots) was lower compared with nucleotide identity of all other genes (white box plots) in all culture conditions (B, batch; C, chemostat; S, starvation). Error bars indicate the standard error of the mean calculated from all genes of all pairwise comparisons. Individual pairwise comparisons are shown in Supplementary Figure S6. ***P«0.01 (student t-test).
Distances between strains are dependent on culture condition and do not follow phylogenetic predictions
Calculated Euclidean distances based on gene expression profiles (gene expression distance (GE distance); see Materials and methods) considering commonly shared genes between the four strains varied considerably between states, where strains showed closest relationship in batch culture, followed by chemostat and starvation (Figure 3a). GE distances did not strictly follow phylogenetic predictions where the degree of difference as well as the rank-order, that is, ranking of pairs based on their phylogenetic relatedness, deviated from ANI values (Figure 3b). The environmental pair displayed lowest GE distances in all culture conditions followed by the laboratory/commensal comparisons, which exhibited 12.3% (median of all comparisons based on commonly shared genes) higher distance values. Similar patterns were obtained for analyses based on all genes, except for batch culture, where GE distances between IAI1 and TW11588 were lower than between environmental strains (Supplementary Figure S7, S8). When only E. coli core genes were considered, distances for environmentals were higher than the laboratory/commensal pair in that state.
Gene expression profile differences (GE distances) between strains in all culture conditions (a) and relative to phylogenetic relatedness (b). All pairwise comparisons based on all commonly shared genes for all culture conditions are shown, where black squares indicate the environmental pair, gray squares display values of the laboratory/commensal pair and open squares show all other comparisons. Black lines indicate mean values of all comparisons (a) and the percentage of the reciprocal ANI (b), respectively. Detailed results based on all genes and E. coli core genes are shown in Supplementary Figures S7 and S8. Differences were significant between batch and chemostat culture (a, P<0.05) and starvation (b, P«0.01). Panel (a) analyses based on Student-Newman-Keuls post-hoc repeated measures ANOVA analysis.
Most RAST gene categories contributed to the observed culture condition-specific GE distances presented in Figure 4. In chemostat culture, 19 categories (total of 24) expressed higher GE distances compared with batch culture, whereas largest GE distances were observed during starvation for 21 categories. Categories associated with metabolism of inorganic substrates namely iron, phosphor, potassium and sulfur displayed largest GE distances in all conditions, whereas the category ‘mobility and chemotaxis’ showed a unique pattern with largest GE distances in batch followed by chemostat and starvation. The latter finding was mainly due to MG1655, which highly expressed flagellum-associated genes, particularly during batch and chemostat growth, whereas these genes were constitutively expressed at low levels in other strains. Gene sets not commonly shared between all tested isolates, referred to as gene-content differences, also contributed significantly to GE distances between strains in both growth states—batch and chemostat; GE-values increased by 54.7% and 34.2%, respectively, when all genes were considered for analysis compared with results based on commonly shared genes only (Supplementary Figure S7). This was not the case in starvation.
Average gene expression distances (GE distance) for individual RAST categories for all culture conditions are displayed (batch, black dots; chemostat, gray triangles; starvation, white squares). The mean and standard deviation of pairwise comparisons of all strains based on all commonly shared genes are given. The numbers indicate the mean of two data points that were above the maximum of the y-axis. AA, amino acids and derivatives; CHO, carbohydrates; Clust, clustering-based subsystems; Co&Vit, cofactors and vitamins; Div&Cyc, cell division and cell cycle; DNA, DNA metabolism; FA&Lip, Fatty Acids and Lipids and isoprenoids; Iron, iron acquisition and metabolism; K-Met, potassium metabolism; Misc, miscellaneous; Mot, motility and chemotaxis; N-Met, nitrogen metabolism; MTrans, membrane transport; N/A, unknown; NsNt, nucleosides and nucleotides; pigments; P-Met, phosphorus metabolism; prosthetic groups; Prot, protein metabolism; Resp, respiration; Rg&Sg, regulation and cell signaling; RNA, RNA metabolism; S-Met, sulfur metabolism; Stress, stress response; Vir&Dis, virulence and disease and defense; W&C, cell wall and capsule. Categories containing ⩾20 genes were included into analysis.
Clustering analysis of gene expression profiles revealed a consistent pattern for chemostat and starvation, where environmental strains grouped together and distinct from the laboratory/commensal strains (Figure 5), whereas this pattern was less conserved for batch cultures. The same result was also observed when only commonly shared genes or E. coli core genes were considered for analysis and shows a robust distinction in overall gene expression profiles between those two pairs (in chemostat and starvation). These finding reveal that the environmental strains follow a similar ecological strategy (despite their distant phylogenetic relation) that is different from the laboratory and enteric model strains. Subsequently, more detailed analysis was performed, where environmental strains and the commensal/laboratory strains, were grouped together, to specifically reveal features that are distinctive between these two pairs.
Gene expression profiles showed a consistent pattern, where environmental strains and laboratory/commensal strains clustered together in all culture conditions. Individual strains are indicated as black-MG: MG1655, dark gray-IAI: IAI1, light gray-T1: TW11588 and white-T2: TW09308. Individual culture conditions, batch (a, d, g), chemostat (b, e, h) and starvation (c, f, i), are shown. Either all genes (a–c) or all commonly shared genes (d–f) or E. coli core genes (g–i) were included in analysis. Replicates are indicated. Bootstrapping of results (100 replicates) revealed absolute consistency (100%) for all plots, except for batch culture when shared genes and E. coli core genes were considered for analysis, where displayed topologies exhibited 83% and 51% consistency, respectively.
Detailed analysis on distinct ecological features between strains
Individual gene (RAST) categories presented in Supplementary Figure S9 follow the overall trend presented in Figure 3a: GE distances within environmentals and the commensal/laboratory strains were consistently lower than between them and many categories clustered these two pairs together (Supplementary Figure S9). The number of upregulated genes for individual pairs was distinct throughout categories in batch and chemostat culture. In starvation, categories associated with growth such as ‘amino acids and derivatives’, ‘carbohydrates’, ‘DNA metabolism’, ‘protein metabolism’, ‘nucleotides/nucleosides’ and others were upregulated in enteric/laboratory strains. In contrast, genes associated with carbon substrate uptake/catabolism and stress response were increasingly expressed in environmental strains in chemostat and starvation (Supplementary Figure S5). In particular genes associated with acid stress defense, toxic compound stress defense, desiccation/osmotic stress defense and the global regulator rpoE were more expressed/induced (compared with batch growth) in environmental strains. In addition, expression of genes responsible for ion balance such as phosphate and potassium transporters was increased in environmental strains during starvation (Supplementary Data set S1). Although expression patterns for the uptake/catabolism of carbon substrates were very similar in all strains, specific differences were also observed, indicating ecological differentiation. For instance, environmental strains showed higher expression of genes linked to threonine and putrescine metabolism in all cultures as well as for fructose (only starvation; Supplementary Figure S5) together with several other, putative, high-affinity sugars/amino acid uptake systems (Supplementary Data set S1). In contrast, enteric/laboratory strains exhibited increased expression of genes associated with uptake/catabolism of several substrates in starvation including fucose, arabinose, ribose, glycerol, acetic acid, fumarate, malate, C4-fatty acids, propionic acid, L-alanyl-L-glutamine/L-alanyl-L-threonine, glycyl-L-aspartic acid and valine. MG1655 displayed a unique pattern with high expression of genes linked to dulcitol, glycerol, glycerolphosphate and oligopeptides in all states. Analysis of group-specific genes (according to the study by Luo et al. (2011)) revealed a few highly expressed genes such as the fucose uptake/catabolism genes, as well as several transcription regulators in enteric/laboratory strains and cell-wall synthesis genes associated with R3 type oligosaccharides in environmental strains.
Clustering of all samples revealed some ‘overlaps’ of strains under starvation, that is, the gene expression profile of a strain was more similar to other strains in starvation than to itself in other states (Figure 6). This phenomenon was more pronounced for commonly shared and core gene sets where overlaps were detected for all states.
Similarity in gene expression profiles of the strains under the same versus different growth conditions. Overall clustering of gene expression profiles showed overlaps between individual strains, that is, the gene expression profile of a strain was more similar to other strains in the same culture condition than to itself in other conditions. Profiles of all strains in all states considering either all genes (a) or shared genes (b) or E. coli core genes (c) are displayed. Strains are represented by color (black: MG1655, dark gray: IAI1, light gray: TW11588 and white: TW09308), whereas letters refer to culture conditions (B, batch; C, chemostat; S, starvation). Replicates and bootstrapped values (⩾80%) are indicated.
Discussion
General aspects of gene expression, growth and ecology
Any given habitat does not dictate a constant behavior in bacteria, but is characterized by many intermittent conditions triggering distinct microbial responses. The ability of E. coli to deal with such (frequently) changing environmental conditions is also reflected in our gene expression data, where the majority of genes were markedly expressed in all culture conditions (Supplementary Figure S2) and is consistent with previous observations for E. coli and other taxa (Passalacqua et al., 2009; Oliver et al., 2009; Haas et al., 2012). The constitutive, though low, expression of most features provides steady templates for translation and we suspect that this enables quick adaptation to changing environmental conditions. Furthermore, microbes often do not adapt to each individual environmental stimulus they encounter with only simple and narrow-scoped responses, but with more general and major gene expression shifts (that is, physiological state), triggered by global regulators such as cAMP, ppGpp and rpoS (Perlman and Pastan, 1969; Lange and Hengge-Aronis, 1991). One intriguing example of global response is the derepression/autoinduction of transporters of many carbon substrates not present in the medium during unlimited growth as observed in our expression data (Supplementary Figure S4) and previous studies (Ihssen and Egli, 2005), supporting this response as a universal feature of E. coli, and probably other heterotrophic bacteria (Egli, 1995). Stress response is another prominent example of global bacterial regulatory mechanisms, where a handful of regulators control expression of many diverse genes generating a phenotype that is cross-protected from many different stresses, and not only from the initial trigger stimulus (Nyström, 2004). This feature appeared to be a key discriminator for E. coli strains from different habitats in our study (Supplementary Figure S5).
Our results emphasize the relative nature of comparative gene-expression investigations, where the culture condition enormously influences results for both within and between strain comparisons. Thus, a controlled experimental set-up is crucial to minimize ‘diverting factors’ such as variation in growth rates between strains to enable the extraction of ecologically relevant signatures. We specifically chose defined, substantially distinct culture conditions to get broad insights into gene-expression profiles of individual strains for different physiological states. Our experimental set-up provided good replication (see for example, Figure 5) and, hence, allowed investigating differential gene expression in detail. We used a single carbon source (glucose) in this study as complex media are ‘unstable’, especially in batch culture where bacteria consecutively feed on more easily degradable substrates. Therefore, we minimized possible biases on gene-expression differences between strains based on distinct substrate preferences (Kovárová-Kovar and Egli, 1998).
Phylogeny, gene regulation and gene-content
We show that gene regulation is directly coupled to gene sequence evolution in E. coli, that is, highly differentially expressed genes show increased coding-sequence dissimilarity (Figure 2). This observation suggests a synergistic effect of these two levels providing a powerful mechanism for fast evolution, consistent with what was recently proposed as an explanation for extreme adaptation in eukaryotes (Castoe et al., 2013). However, the link between phylogeny and gene regulation is not always tightly linked as actual GE-distance calculations did not strictly follow ANI relatedness, demonstrating that gene regulation presents an important additional mechanism for diversification in E. coli. Overall, our results suggest that gene regulation of commonly shared genes (and E. coli core genes), rather than gene-content differences, might be driving diversification as previously suggested (Phillipe et al., 2007); at least in frequently encountered conditions such as starvation or carbon-limited growth. Although, non-commonly shared genes (reflecting differences in gene-content) significantly added to distances between strains and buffered some of the overlaps in gene expression profiles (Figure 6), they played a relatively minor role for lineage specific distinction, at least for the physiological states investigated here. For instance, clustering results presented in Figure 4 are robust based on commonly shared genes and even E. coli core genes only (not for batch). Furthermore, the majority of non-commonly shared genes were expressed at low levels in all culture conditions, questioning their functionality in vivo. However, the possibility that those genes do not fall under the regulatory mechanisms governing the physiological states investigated here, but are only expressed under specific circumstances not covered in this study, remains.
GE distances between strains depend on the physiological state
The observed differences in GE distances between culture conditions demonstrate that relationship between bacteria are not constant, but depend on the physiological state. Accordingly, smallest GE distances were observed in exponentially growing batch cultures, resulting presumably from elevated purifying selection processes during growth in that state, where the main objective for members of all lineages is fast multiplication. In contrast, other features, such as growth efficiency or stress resistance are less important under exponential growth. Furthermore, certain growth conditions trigger broader responses than others such as the derepression/autoinduction of many carbon source pathways in chemostat culture, which contributed to the observed GE distances. The larger GE distances measured in starvation relative to growth conditions are probably a combination of expression patterns specifically evolved in that state, and a direct result of distinct strain physiologies in the former state, in our case carbon-limited growth, which additionally directed responses when strains were entering starvation.
Variations in GE (and eventually phenotypic) distances can have direct ecological implications. For example, mixed substrate growth conditions can reduce niche overlaps between bacteria, enabling a higher capacity for diversity by creating large (expression) distances between taxa. In other words, bacteria growing in an environment where a carbon source that initiates catabolic repression is abundant (for example, glucose) show tighter substrate niche overlaps (as secondary substrates are neglected) compared with conditions where mixed substrate growth prevails (for example, carbon-limited or arabinose-rich environment (Ihssen and Egli, 2005). Additional experiments specifically focusing on the influence of the (GE)distance on bacterial competition/selection will contribute to this field in future.
Signatures of ecological adaptation
The small distances and consistent grouping of the two environmentals in all culture conditions, together with differences in their ecological strategy compared with other strains, suggest that they do indeed form a separate ecotype as proposed earlier, where extra-intestinal open environments serve as their primary habitat (Walk et al., 2009; Luo et al., 2011). It should be noted that E. coli strains belonging to the same clades as the strains used in this study are commonly detected in bird feces as well (Clermont et al., 2011), but it is currently not clear if these strains cycle between birds and the environment or whether different ecotypes with distinct primary habitats do exist within those lineages. Regardless of this lack of complete understanding of the breadth of the ecological niche of the strains, Ecotype Simulations and AdaptML analysis from the study by Cohan and Kopac indicated that atypical strains from lineage IV do form a distinct ecotype to members of clade V (Cohan and Kopac, 2011). Specific comparisons of, for example, gene expression profiles (as done in this study) or specific growth properties (c.f. Vital et al., 2012) between strains that were isolated from both the environment and birds could add clarification on this issue.
Two distinct ecological strategies were observed. Environmental strains expressed numerous stress defense genes at higher levels than other strains, already during carbon-limited growth, whereas commensal/laboratory strains retain many genes associated with carbon substrate catabolism and central metabolism upregulated in starvation. These findings suggest that environmental strains are more prepared for no-growth conditions, which allows them to cope with starvation more quickly. Thus, a trade-off in investing energy for staying alert for nutrient scavenging/assimilation during non-growth conditions versus entering a survival phenotype seems to be a major feature of distinction between strains and reflects well their preferred habitat: non-growth states are less frequent and are much shorter in the laboratory and the human gut, whereas starvation conditions prevail in the environment, rendering energy expenses for the expression of surviving features advantageous. This trade-off was also proposed as an explanation for the high polymorphism in the rpoS gene, where specifically enteric strains have attenuated phenotypes or are completely lacking a functional gene (Ferenci, 2003). Rapid displacement of wild-type strains by strains exhibiting weaker stress responses (for example, rpoS mutants) is also commonly observed in the laboratory (Notley-McRobb et al., 2002). In general, while MG1655 displayed unique features that might be specifically associated with its primary habitat (the laboratory), such as high expression of flagellum-associated genes, the strain consistently clustered together with the commensal and displayed an overall similar strategy validating the human gut as its origin. However, the distances between the two strains were consistently larger than those between environmental strains (despite their closer phylogenetic relationship), which suggests partly distinct ecological adaptations via gene-regulatory mechanisms in MG1655.
Affinities to certain substrates characteristic for the individual habitats such as fucose, a common component of host mucus glycans (Chang et al., 2004), or the plant-derived putrescine (Flores and Galston, 1982) also contributed to ecology-specific gene expression profiles. Furthermore, the high expression of phosphorus uptake systems in environmental strains during starvation might stem from recurrent dual-carbon-/phosphorus-limited conditions specifically in their habitat, which led to an increased co-expression of this system during carbon limitation (Zinn et al., 2004). It should also be noted that numerous highly expressed genes are of unknown function for both commonly shared genes and group-specific genes and their potential role in niche adaptation remains elusive. While this study provides the first quantitative data on the importance of gene expression for defining lineage-diagnostic signatures and identifies habitat-specific gene regulation and activity, only a few strains were examined. Hence, it provides hypotheses on ecotype-specific markers to be tested with additional strains under different culture conditions.
Overlapping expression profiles
We observed that signatures of the physiological state were often more pronounced in gene expression profiles than phylogenetic strain boundaries, in particular for commonly shared features (Figure 6). This implies that all E. coli strains evolve similarly under a specific growth condition, suggesting that divergence rates are not constant over time, but amplify once an ecological barrier is crossed to a new habitat where distinct growth conditions prevail. Trade-offs between adaptation to/within a specific habitat at the expense of optimal performance in other environments is recognized in E. coli (Ferenci, 2003). Finally, while the individual analysis in each state justifies the environmental E. coli as a separate species from human enteric models, the overall ‘expression pattern overlaps’ (Figure 6) indicate incomplete divergence. However, it is currently unclear what level of gene expression differences should translate to (new) species descriptions and how established methods for delineating taxonomic ranks, especially at the species level, correspond to gene expression profile differences or similarities. Our study provides the first insight into this field and a guide for future investigations, including additional assays such as proteomics/enzymatic analysis on different taxa to investigate the significance of observed overlaps for bacterial speciation.
References
Anders S, Huber W . (2010). Differential expression analysis for sequence count data. Genome Biol 11: R106.
Caro-Quintero A, Konstantinidis KT . (2012). Bacterial species may exist, metagenomics reveal. Environ Microbiol 14: 347–355.
Castoe T, de Koning PJ, Hall KT, Card DC, Schield DR, Fujita MK et al. (2013). The Burmese python genome reveals the molecular basis for extreme adaptation in snakes. Proc Natl Acad Sci USA 110: 1–6.
Chang DE, Smalley DJ, Tucker DL, Leatham MP, Norris WE, Stevenson SJ et al. (2004). Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci USA 101: 7427–7432.
Clermont O, Gordon DM, Brisse S, Walk ST, Denamur E . (2011). Characterization of the cryptic Escherichia lineages: rapid identification and prevalence. Environ Microbiol 13: 2468–2477.
Cohan FM, Kopac SM . (2011). Microbial genomics: E. coli relatives out of doors and out of body. Curr Biol 21: 587–589.
Egli T . (1995). The ecological and physiological significance of the growth of heterotrophic microorganisms with mixtures of substrates. Adv Microb Ecol 14: 305–386.
Enard W, Khaitovich P, Klose J, Zöllner S, Heissig F, Giavalisco P et al. (2002). Intra- and interspecific variation in primate gene expression patterns. Science 296: 340–343.
Ferenci T . (2003). What is driving the acquisition of mutS and rpoS polymorphisms in Escherichia coli? Trends Microbiol 11: 457–461.
Flores HE, Galston AW . (1982). Analysis of polyamines in higher plants by high performance liquid chromatography. Plant Physiol 69: 701–706.
Goris J, Konstantinidis KT, Klappenbach J, Coenye T, Vandamme P, Tiedje JM . (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57: 81–91.
Haas BJ, Chin M, Nusbaum C, Birren BW, Livny J . (2012). How deep is deep enough for RNA-Seq profiling of bacterial transcriptomes? BMC Genomics 13: 734.
Huwiler SG, Beyer C, Fröhlich J, Hennecke H, Egli T, Schürmann D et al. (2012). Genome-wide transcription analysis of Escherichia coli in response to extremely low-frequency magnetic fields. Bioelectromagnetics 33: 488–496.
Ihssen J, Egli T . (2005). Global physiological analysis of carbon- and energy-limited growing Escherichia coli confirms a high degree of catabolic flexibility and preparedness for mixed substrate utilization. Environ Microbiol 7: 1568–1581.
Ihssen J, Egli T . (2004). Specific growth rate and not cell density controls the general stress response in Escherichia coli. Microbiology 150: 1637–1648.
Ihssen J, Grasselli E, Bassin C, François P, Piffaretti JC, Köster W et al. (2007). Comparative genomic hybridization and physiological characterization of environmental isolates indicate that significant (eco-)physiological properties are highly conserved in the species Escherichia coli. Microbiology 153: 2052–2066.
King M, Wilson A . (1975). Evolution at two levels in humans and chimpanzees. Science 188: 107–116.
Konstantinidis KT, Ramette A, Tiedje JM . (2006). The bacterial species definition in the genomic era. Philos Trans R Soc Lond B Biol Sci 361: 1929–1940.
Konstantinidis KT, Serres MH, Romine MF, Rodrigues JLM, Auchtung J, McCue LA et al. (2009). Comparative systems biology across an evolutionary gradient within the Shewanella genus. Proc Natl Acad Sci USA 106: 15909–15914.
Kovárová-Kovar K, Egli T . (1998). Growth kinetics of suspended microbial cells: from single-substrate-controlled growth to mixed-substrate kinetics. Microbiol Mol Biol 62: 646–666.
Lange R, Hengge-Aronis R . (1991). Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol 5: 49–59.
Langmead B, Trapnell C, Pop M, Salzberg SL . (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25.
Luo C, Walk ST, Gordon DM, Feldgarden M, Tiedje JM, Konstantinidis KT . (2011). Genome sequencing of environmental Escherichia coli expands understanding of the ecology and speciation of the model bacterial species. Proc Natl Acad Sci USA 108: 7200–7205.
Notley-McRobb L, King T, Ferenci T . (2002). rpoS mutations and loss of general stress resistance in Escherichia coli populations as a consequence of conflict between competing stress responses. J Bacteriol 184: 806–811.
Nyström T . (2004). Stationary-phase physiology. Annu Rev Microbiol 58: 161–181.
Oliver HF, Orsi RH, Ponnala L, Keich U, Wang W, Sun Q et al. (2009). Deep RNA sequencing of L. monocytogenes reveals overlapping and extensive stationary phase and sigma B-dependent transcriptomes, including multiple highly transcribed noncoding RNAs. BMC Genomics 10: 641.
Passalacqua KD, Varadarajan A, Ondov BD, Okou DT, Zwick ME, Bergman NH . (2009). Structure and complexity of a bacterial transcriptome. J Bacteriol 191: 3203–3211.
Perlman R, Pastan I . (1969). Pleiotropic deficiency of carbohydrate utilization in an adenyl cyclase deficient mutant of Escherichia coli. Biochem Biophys Res Commun 37: 151–157.
Philippe N, Crozat E, Lenski RE, Schneider D . (2007). Evolution of global regulatory networks during a long-term experiment with Escherichia coli. Bioessays 29: 846–860.
Rosselló-Mora R, Amann R . (2001). The species concept for prokaryotes. FEMS Microbiol Rev 25: 39–67.
Stern DL, Orgogozo V . (2008). The loci of evolution: how predictable is genetic evolution? Evolution 62: 2155–2177.
Van Valen L . (1976). Ecological species, multispecies, and oaks. Taxon 25: 233–239.
Vital M, Hammes F, Egli T . (2012). Competition of Escherichia coli O157 with a drinking water bacterial community at low nutrient concentrations. Water Res 46: 6279–6290.
Walk ST, Alm EW, Gordon DM, Ram JL, Toranzos G, Tiedje JM et al. (2009). Cryptic lineages of the genus Escherichia. Appl Environ Microbiol 75: 6534–6544.
Zinn M, Witholt B, Egli T . (2004). Dual nutrient limited growth: models, experimental observations, and applications. J Biotechnol 113: 263–279.
Acknowledgements
We thank Professor Denamur for providing the IAI1 strain and Chengwei (Alex) Luo for the E. coli pangenome table. Financial support was provided in part by NIH Human Microbiome Project Demonstration Project (UH3 DK083993) and by NSF DEB-0516252. Microbial informatics support was provided by RDP under grant DE-FG02-98ER62848 from the Office of Science, US Department of Energy.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Vital, M., Chai, B., Østman, B. et al. Gene expression analysis of E. coli strains provides insights into the role of gene regulation in diversification. ISME J 9, 1130–1140 (2015). https://doi.org/10.1038/ismej.2014.204
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2014.204
This article is cited by
-
Glucose consumption rate-dependent transcriptome profiling of Escherichia coli provides insight on performance as microbial factories
Microbial Cell Factories (2022)
-
Synergistic cellulose-based nanocomposite packaging and cold plasma decontamination for extended saffron preservation
Scientific Reports (2022)
-
The challenges in the identification of Escherichia coli from environmental samples and their genetic characterization
Environmental Science and Pollution Research (2022)
-
Phylogenomic Analyses of a Clade Within the Family Flavobacteriaceae Suggest Taxonomic Reassignments of Species of the Genera Algibacter, Hyunsoonleella, Jejuia, and Flavivirga, and the Proposal of Pseudalgibacter gen. nov. and Pseudalgibacter alginicilyticus comb. nov.
Current Microbiology (2021)
-
Reclassification of Algibacter wandonensis as a Later Heterotypic Synonym of Algibacter lectus Based on Whole-Genome Sequence Analysis
Current Microbiology (2021)