Abstract
A seven-year oceanographic time series in NW Mediterranean surface waters was combined with pyrosequencing of ribosomal RNA (16S rRNA) and ribosomal RNA gene copies (16S rDNA) to examine the environmental controls on SAR11 ecotype dynamics and potential activity. SAR11 diversity exhibited pronounced seasonal cycles remarkably similar to total bacterial diversity. The timing of diversity maxima was similar across narrow and broad phylogenetic clades and strongly associated with deep winter mixing. Diversity minima were associated with periods of stratification that were low in nutrients and phytoplankton biomass and characterised by intense phosphate limitation (turnover time<5 h). We propose a conceptual framework in which physical mixing of the water column periodically resets SAR11 communities to a high diversity state and the seasonal evolution of phosphate limitation competitively excludes deeper-dwelling ecotypes to promote low diversity states dominated (>80%) by SAR11 Ia. A partial least squares (PLS) regression model was developed that could reliably predict sequence abundances of SAR11 ecotypes (Q2=0.70) from measured environmental variables, of which mixed layer depth was quantitatively the most important. Comparison of clade-level SAR11 rRNA:rDNA signals with leucine incorporation enabled us to partially validate the use of these ratios as an in-situ activity measure. However, temporal trends in the activity of SAR11 ecotypes and their relationship to environmental variables were unclear. The strong and predictable temporal patterns observed in SAR11 sequence abundance was not linked to metabolic activity of different ecotypes at the phylogenetic and temporal resolution of our study.
Similar content being viewed by others
Introduction
An important goal in microbial oceanography is to link the extensive diversity of marine bacteria (Venter et al., 2004; Sogin et al., 2006) to environmental conditions (Morris et al., 2005; Fuhrman et al., 2006; Gilbert et al., 2012), ecosystem function (Fuhrman, 2009; Galdos et al., 2011) and climate sensitivity (Eiler et al., 2009; Giovannoni and Vergin, 2012). Combined time-series of microbial community diversity and oceanographic parameters have emerged as a useful approach to understand how prokaryotic communities are distributed across ecological niches (Carlson et al., 2009; Hugoni et al., 2013). Temporal dynamics of marine bacteria over short timescales are reasonably well studied and strong seasonal transitions in community structure are evident across different environments (Alonso-Sáez et al., 2007; Campbell et al., 2009; Lami et al., 2009). However, there are few extended microbial time-series (>4 years), which are necessary to establish predictable and statistically robust patterns in microbial community dynamics.
The long-term microbial studies conducted to date have revealed annually recurring patterns of surface bacterial communities. In Californian coastal waters these patterns can be reliably predicted from seasonality in environmental parameters (Fuhrman et al., 2006; Chow et al., 2013). In the Sargasso Sea bacterial dynamics exhibit a strong seasonal component that appears to be driven by convective overturning (Morris et al., 2005; Carlson et al., 2009; Treusch et al., 2009). Repeatable seasonality in bacterial diversity is also evident from the Western English Channel (Gilbert et al., 2012) and appears to be a general feature common of mid- to high-latitude environments (Ladau et al., 2013). Although seasonal cycles of bacterial diversity are an emerging paradigm in microbial oceanography, the factors controlling these cycles and their significance for ecosystem function are poorly understood. Furthermore, it is unknown whether seasonality in the metabolic activity of different bacterioplankton clades follow similar cycles.
Estimating the metabolic activity of phylogenetically defined bacteria from mixed communities is challenging. Ribosomal RNA (rRNA) is commonly used to characterise active microbial populations (Troussellier et al., 2002; Gentile et al., 2006) and the frequently observed discrepancies in profiles of rRNA and ribosomal RNA genes (rDNA; Moessender et al., 2001; Lami et al., 2009) have been interpreted to represent different metabolic states (Treusch et al., 2009; Brettar et al., 2011). The basis of this approach is the observation that cell ribosome content increases with growth rate (DeLong et al., 1989; Fegatella et al., 1998; Kerkhoff and Kemp, 1999). More recent application of this principle has used rRNA/rDNA ratios as an in-situ proxy for growth/activity rates (Campbell et al., 2011; Campbell and Kirchman, 2013; Hunt et al., 2013).
Owing to the association of particular metabolic activities to certain clades, changes in the activity of specific, and in particular dominant, bacterial communities have the potential to affect ocean carbon and nutrient cycling. In this context the SAR11 clade of the Alphaproteobacteria is of particular interest because it is a dominant component of marine bacteria (Morris et al., 2002). Ubiquitously distributed throughout the ocean (Giovannoni and Rappé, 2000) SAR11 typically accounts for 15–35% of all prokaryotic plankton (Malmstrom et al., 2005, 2007; Lefort and Gasol, 2013). Furthermore, SAR11 are known to take up a variety of monomeric substrates including amino acids, glucose, ATP and dimethylsulfoniopropionate (Malmstrom et al., 2004; Alonso-Sáez and Gasol, 2007; Laghdass et al., 2012), and appears to be an important organism for a variety of biogeochemical functions. In oligotrophic environments such as the NW Mediterranean Sea, SAR11 can dominate bacterial production. Contributions towards total leucine assimilation may reach 60–70% in nutrient-limited summer months (Laghdass et al., 2010, 2012). The apparent importance of SAR11 to ecosystem function in oligotrophic environments may stem from adaptations to low substrate conditions: a streamlined genome (Giovannoni et al., 2005a; Grote et al., 2012) and the use of light-harvesting proteorhodopsins (Giovannoni et al., 2005b) may convey significant ecological advantages.
Despite these important generalisations concerning the abundance and activity of SAR11 as a clade, it is a diverse group (Field et al., 1997; Garcia-Martinez and Rodriguez-Valera, 2000; Suzuki et al., 2001; Brown and Fuhrman, 2005) comprised of multiple sub-clades (Morris et al., 2005; Rusch et al., 2007; Brown et al., 2012). 16S rRNA gene sequencing and molecular fingerprinting approaches have shown SAR11 sub-clades to be partitioned by season, depth and biome (Field et al., 1997; Garcia-Martinez and Rodriguez-Valera, 2000; Stingl et al., 2007; Carlson et al., 2009; Beman et al., 2011; Brown et al., 2012). This ecological niche separation of SAR11 sub-groups Ia, Ib and II, led to their classification as ecotype-like clades, with potential for fine-scale phylogenetic structure within these groups (Vergin et al., 2013a). The clear temporal and spatial patterns observed among SAR11 communities are thought to result from the variable response of specific ecotypes to different environmental conditions (Beman et al., 2011; Carlson et al., 2009; Brown et al., 2012). However, there are no direct attempts to link seasonal trends in SAR11 ecotype abundance and activity over an extended microbial time-series. To understand the role of these clades in microbial community assembly and ecosystem function (Koeppel et al., 2008) it is necessary to differentiate patterns of abundance and activity in SAR11 ecotypes.
In the present study we sequenced rRNA genes (named rDNA hereafter) and rRNA of surface bacterial communities from a 7-year time-series at monthly resolution in the NW Mediterranean. Short-term (<2 years) studies in coastal waters of the NW Mediterranean have shown important seasonal transitions among bacterial communities and general significance of the entire SAR11 clade (Schauer et al., 2003; Alonso-Sáez et al., 2007; Lami et al., 2009). The aim of the current study was to characterise the seasonal dynamics of SAR11 diversity and ecotype relative abundance and activity at an offshore site. Our main objectives were to determine (i) if total bacterial diversity and that within the SAR11 clade varies seasonally (ii) if dominant ecotypes show specific seasonal patterns in relative abundance and (iii) whether certain biogeochemical parameters constrain ecotype abundance and activity.
Materials and methods
Sampling
Surface seawater (3–5 m) was collected at the Microbial Observatory of the Laboratoire Arago located at 47°27′136 N, 03°32′360 E in the North-West Mediterranean. Samples were collected approximately every month from July 2004 to April 2011. Occasionally samples were not collected due to poor weather conditions. Full sampling details are given in Supplementary Information and exact sampling dates are summarised in Supplementary Table 1. Depth profiles of conductivity, temperature and pressure were obtained with a Sea Bird SBE 19 profiler.
Chemical and physical data
Nutrients
Concentrations of nitrate, nitrite, phosphate and silicate were measured with a Skalar auto-analyser following a previously described protocol (Treguer and Le Corre, 1975).
Ammonium
Dissolved ammonium was measured by spectrophotometry at 630 nm following conversion to indophenol via a monochloroamine intermediate (Solorzano, 1969).
Chlorophyll
One litre of seawater was collected on a GF/F filter at low pressure (<0.2 bar). Measurements were conducted according to previously published protocols (Yentsch and Menzel, 1963; Strickland and Parsons, 1997). Extended protocol details can be found in Supplementary Information.
Mixed layer depth (MLD)
Raw seabird data were processed to bin temperature and salinity data at 0.25 m intervals. MLD is based on a 0.1 °C and/or 0.02-sigma difference from a reference threshold (typically an average of the upper 3 m).
Microbial abundances
Unfiltered seawater samples were fixed at a final concentration of 1% glutaraldehyde, incubated for 15 min at ambient temperature in the dark, frozen in liquid nitrogen and stored at −80 °C. Cytometry analyses were performed on a Becton Dickinson FacsCalibur. Full details of preparation and analysis of cytometry samples can be found in Supplementary Information.
Microbial rate measurements
Bacterial heterotrophic production
The incorporation of 3H-leucince was measured using the centrifugation method (Smith and Azam, 1992; Kirchman, 1993). Extended protocol details can be found in Supplementary Information.
SAR11 bacterial production
To determine bacterial production by SAR11 cells, fluorescence in-situ hybridisation was coupled with microautoradiography (MICRO-CARD-FISH). Identical to measurements for bulk bacteria, tritiated leucine was used as a precursor for SAR11 bacterial production (for example, Laghdass et al., 2012). Extended protocol details can be found in Supplementary Information.
Phosphate turnover
These measurements were performed during 2012 at a location in the NW Mediterranean in close proximity (42°31′N, 03°11′E) to station Microbial Observatory of the Laboratoire Arago (47°27′N, 03°32′E). The turnover of phosphate by microbial populations was determined using a concentration series bioassay (Wright and Hobbie, 1966; Zubkov et al., 2007). Extended protocol details can be found in Supplementary Information.
Nucleic acid extraction, reverse transcription-PCR and pyrosequencing
The extraction of nucleic acids (DNA and RNA) was performed according to a previously published method (West et al., 2008). A combination of mechanic (freeze-thaw) and enzymatic cell lysis techniques were applied directly to Sterivex cartridges, followed by extraction using an AllPrep DNA/RNA kit (Qiagen, Venlo, Netherlands). The RNA samples were tested for the presence of contaminating genomic DNA by PCR and then reverse-transcribed with random primers using the SuperScript III Reverse Transcriptase kit (Invitrogen, Carlsbad, CA, USA). We are unable to account for potential biases associated with reverse transcription efficiency and RNA secondary structure within the present data set. The amplification of the V1–V3 region of the 16S rRNA gene was performed with universal bacterial primers 28F (5′-TTTGATCNTGGCTCAG-3′) and 519R (5′-GTNTTACNGCGGCKGCTG-3′), followed in pyrosequencing in a commercial laboratory (Research and Testing Laboratory, Lubbock, TX, USA) using a Roche 454 GS-FLX system (Roche, Basel, Switzerland) with titanium chemistry. All sequences have been submitted to the sequence read archive under the Bioproject accession number: PRJNA235253.
Sequence data analyses
Sequences were analysed as previously described (Blanquer et al., 2013). Briefly, the reads were first filtered by removing low quality reads and subsequently trimmed to remove those with ⩾3% of bases with Phred values <27 (0.2% per-base error probability). This is recommended to ensure that when clustering at 97% the influence of erroneous reads is minimised (Huse et al., 2010; Kunin et al., 2010). Sequences were then clustered at a 97% threshold using Uclust algorithm (Edgar, 2010) and each operational taxonomic unit (OTU) classified by comparison to the Greengenes database (DeSantis et al., 2006). Sequence analyses were conducted with Pyrotagger (Kunin and Hugenholtz, 2010).
A phylogenetic tree was constructed with the sequence representatives of the abundant SAR11 OTUs (<0.2%). Sequences included in the tree were deposited in GenBank under accession numbers KJ094527–KJ094552. Representative sequences for each OTU together were aligned using MUSCLE (Edgar, 2004) and phylogenetic analyses were completed with the programme PHYLIP (Felsenstein, 2008). DNADIST was used to calculate genetic distances using a Kimura-2 model. The distance tree was estimated with the phylogenetic algorithm FITCH in the PHYLIP programme.
To compare bacterial communities for diversity analysis, all samples were randomly re-sampled to the size of the sample containing fewest sequences (n=446) using Daisy Chopper (Gilbert et al., 2009). The Shannon diversity index (H′) was calculated using the software PAST (Hammer et al., 2001).
Statistical analyses and partial least squares regression (PLSR)
Unpaired t-tests
Samples were grouped into high or low diversity regimes using a constant statistical threshold determined empirically across the 7-year data set. The two categories contained sample populations of 13 and 12, respectively. Average values and standard deviations for each measured environmental parameter were determined and unpaired t-tests were used to calculate statistical differences between individual parameters within the two groups. The null hypothesis (H0) was rejected if the P-value was <0.05 (GraphPad, GraphPad Software Inc, San Diego, CA, USA). A relatively large number of independent comparisons were made and we therefore applied Benjamini–Hochberg False Discovery Rate to calculate an adjusted P-value (Benjamini and Hochberg, 1995), which we denote as P* in Table 1. Results are statistically significant if P*<0.05.
Co-variance (COV)
To determine the nature of COV between the rRNA/rDNA ratios of different ecotypes, the following formula was used:

Where x and y are dependent and independent variables and n is the number of observations.
Values >0 and <0 indicate positive and negative COV, respectively.
PLSR
Models were developed based on projection to latent structures using PLSR. PLSR was selected as a predictive tool over multiple linear regression because the latter is only valid when factors are not significantly redundant (co-linear). PLSR linearly extracts few latent variables that are most useful in modelling the response. We used partial least squares (PLS) to predict multiple Y values (SAR11 ecotype sequence abundances) from a set of X variables (environmental parameters). The model was constructed using a 4-year sub-set (2007–2011) of the 7-year data, for which all environmental parameters were available (Supplementary Table 1). All analyses were performed with UnscramblerX (Camo, Oslo, Norway). Input data were auto-scaled by dividing each x and y value by its standard deviation. We used the Non-Linear Iterative Partial Least Squares algorithm. Cross-validated R2 values (Q2) were taken as a measure of the models Goodness of Fit (Wold et al., 2001). The PLSR model we developed was optimised with a Q2 value of 0.70 at three latent factors (Supplementary Figure 1). Weighted regression coefficients were calculated for each x-variable and SAR11 sequence abundance data set (that is, Y1:4) to ascertain the most important measured environmental variables describing SAR11 ecotype abundance.
Inter-quartile range
Using the inter-quartile range principle [Q75+c(Q75−Q25)], where Q25 and Q75 are lower and upper quartiles and c is a positive constant, one of the μCARD-FISH data points was identified as a potential outlier. This single data point was removed when calculating regression coefficients.
Results
Sequence distributions
We pyrosequenced 57 16S rDNA and 50 16S rRNA samples collected as part of an offshore monthly time-series in the North-West Mediterranean. A total 66 705 16S rDNA and 57 524 rRNA sequences were obtained after performing quality filtering. Sequences of rRNA and rDNA from all sampling dates were pooled and clustered to yield a total of 2222 OTUs. The mean number of sequences obtained per sample was 1170±470 for rDNA and 1150±411 for rRNA. Descriptive statistics summarising the central tendencies and dispersion are provided in Supplementary Table 2. The vast majority of sequences were assigned to either members of the SAR11 clade or classified as members of the Cyanobacteria phylum. Classification of Cyanobacteria into lower taxonomic ranks was not possible using the sequenced region. The contribution of SAR11 to total sequences had medians of 78 and 45% in the rDNA and rRNA fractions compared with 13 and 46% for Cyanobacteria.
Total diversity and empirical identification of diversity regimes
Diversity in each individual sample (Shannon index) was estimated using the rDNA data set after random re-sampling. The Shannon diversity index ranged from 1 to 3.5 and a distinct seasonal cycle in diversity was observed with maxima occurring in the winter and minima in the summer (Figure 1). Diversity within the SAR11 group was calculated in a similar way without re-sampling. The median number of SAR11 sequences in diversity calculations was 344 (−0.68σ=209; +0.68σ=388). The SAR11 Shannon index ranged from 0.03 to 2.3. The number of SAR11 OTUs ranged from 1 to 65.
Seasonal cycles in total and SAR11 diversity. Shannon diversity index is calculated from tag sequence data of ribosomal RNA gene copies (rDNA). The horizontal dashed and dotted lines are empirical thresholds for high and low diversity regimes, which are marked by grey and white triangles, respectively. The dashed blue and solid red lines are SAR11 diversity expressed as Shannon index (H′) and OTU richness (S), respectively.
The observed seasonality in bacterial diversity was used to define an empirical framework of high and low diversity regimes. Individual sampling dates were classified as high or low diversity by comparison to a constant statistical threshold of 2.08±0.55 (arithmetic mean ±1σ). The exception to this is the high diversity peak of 2005/2006, which was slightly below the threshold criterion, nevertheless it was included to ensure at least one representative from the different diversity regimes were available for each sampling year. Partitioning the data in this way identified 25 sampling dates consisting of the 13 highest and 12 lowest estimates of diversity (Figure 1).
Biogeochemical conditions characterising the diversity regimes
Pairwise t-tests were used to determine whether the operationally defined diversity regimes were also segregated based on biogeochemical conditions (Table 1). The most significant differences in biogeochemical parameters among these regimes (P<0.0001) were deeper mixed layers and lower temperatures at times of high diversity. In these samples, nitrate (P=0.001), chlorophyll (P=0.03) and nitrite (P=0.0003) concentrations were significantly higher. The magnitude of the differences and significance levels for environmental comparisons are summarised in Table 1.
SAR11 ecotype distributions
SAR11 Ia ecotypes were the dominant SAR11 clade in all samples and accounted for >60% of total SAR11 rDNA sequences in high and low diversity regimes (Figure 2). Percentages of SAR11 Ib and SAR11 II rDNA increased from 3.3±2.8 and 2.4±2.8 at times of low diversity to 13.4±4.2 and 6.6±3.2 at times of high diversity, but these differences were not statistically significant. When combining relative abundances of clades Ib and II, the differences between high and low diversity regimes were significant and indicated that the temporal dynamics of Ib and II are independent of Ia.
Phylogenetic relationship and relative abundance of SAR11 OTUs. Only SAR11 OTUs representing >0.2% of total SAR11 OTU sequences are included. Bubble size is scaled to sequence abundance of OTUs relative to total SAR11 communities. DNA and RNA is 16S rRNA gene copies and 16S rRNA, respectively. High and low diversity sub-groups were empirically defined; see Figure 1 and text for further details.
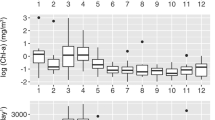
Month to month differences in ecotype distributions were also evident (Figure 3). Both the rDNA and rRNA sequences of ecotype Ia showed maximum abundances of 81–87% from April to October, a period corresponding to higher leucine incorporation (16–42 pmol l−1 h−1) and extremely rapid phosphate turnover times (<5 h, Figure 3a). Ecotypes Ib and II displayed consistently inverse patterns to Ia but were broadly consistent to each other with peaks from November to March. During this period leucine incorporation was lower (5.0–7.4 pmol l−1 Hr−1) and phosphate turnover was longer ranging from 30 to 130 h with highest values found in December and January (Figure 3).
Monthly averages of SAR11 ecotype abundance and microbial rate processes. The monthly averages of sequence abundance are calculated from (a) ribosomal RNA gene copies and (b) ribosomal RNA tag-sequence data. The abundance of sequences in SAR11 ecotype classifications Ia, Ib and II are cumulative sequences within the phylogenetically defined sub-clades (Figure 2) expressed relative to total SAR11 sequences. The difference of SAR11 sequences not included in these sub-clade classifications is represented by the blank area at the top of panels. White, light grey and dark grey areas represent SAR11 Ia, Ib and II, respectively. White circles and solid line in panel a are phosphate turnover times determined in the NW Mediterranean during 2012, error bars are analytical errors on triplicate measurements. White squares and dashed lines in panel b are leucine incorporation rates temporally binned with 1-month resolution. Error bars are standard errors on monthly measurements integrated over the 7-year time-series.
PLSR model
The relative abundance of SAR11 ecotypes from both rDNA and rRNA sequences were combined with biogeochemical observations to develop a model that described SAR11 ecotype dynamics based on PLS regression analysis (Figure 4). The biogeochemical variables included in the model are listed in Table 1. Using a three-factor component model with non-transformed input variables, the model was capable of describing 70% of observed variance in the temporal dynamics of SAR11 ecotype abundance. The goodness of fit for individual ecotype groupings based on rRNA abundances was higher (0.76–0.78) than those based on rDNA abundances (0.63–0.66; Figure 4a). All data were centred and auto-scaled to compute weighted regression coefficients for the biogeochemical input variables (Figure 4b). Regression coefficients were uniformly larger for rRNA abundance data, although similar to rDNA. The physical parameters of MLD, temperature and salinity were quantitatively the most important variables for describing temporal trends of SAR11 ecotype abundance.
Partial least squares regression model of SAR11 ecotype abundance. (a) The relationship between modelled and observed SAR11 ecotype distributions from ribosomal RNA gene copies and ribosomal RNA tag-sequence data. (b) The weighted regression coefficient of each biogeochemical parameter used to model SAR11 ecotype patterns. Positive values indicate that the input variable is positively correlated to specific output variables. MLD, mixed layer depth; Pro., Prochlorococcus; Chl., chlorophyll; NanoEuK., nanoeukaryotes; Bac. Abund., bacterial abundance; Bac. Prod., bacterial productivity; Syn., Synechococcus, PicoEuk., picoeukaryotes, Temp., temperature.
rRNA/rDNA activity ratios
Sequences were grouped to calculate rRNA/rDNA ratios of the SAR11 clade and compared with estimates of SAR11 leucine incorporation (Figure 5). The μCARD-FISH samples were collected as part of a separate project and were only available for the year 2008 (Laghdass et al., 2012). SAR11 leucine incorporation and rRNA/rDNA ratios were positively correlated (R=0.86; P<0.05). The rRNA/rDNA ratios of SAR11 ecotypes were not correlated with respective abundances (Figure 6a). The PLSR modelling approach was unable to capture temporal dynamics of activity ratios with a satisfactory goodness of fit (Supplementary Figure 2). However, rRNA/rDNA ratios of all individual ecotype pairings (Ia vs Ib, Ib vs II, and Ia vs II) exhibited positive covariance (methods) indicating that they were generally active at the same times and their activity ratios were not inversely related (Figure 6). SAR11 ecotype rRNA/rDNA ratios were compared over the range 0–1.5. Data that exceeded 1.5 were atypical (<4%) and not considered to be representative of the general trends within the data set. The largest positive correlation was found between Ia and Ib (R=0.56; P<0.001), followed by Ib and II (R=0.47; P<0.01) and Ia and II (R=0.38; P<0.05).
Leucine incorporation and RNA/DNA ratios in the SAR11 clade. The ratio of ribosomal RNA (rRNA) to ribosomal RNA gene copies (rDNA) were calculated by summing the sequences for all SAR11 clusters. SAR11-active cells were calculated as the percentage of total SAR11 cells assimilating tritiated leucine.
rRNA/rDNA activity ratios and abundance of SAR11 ecotypes. (a) The relationship between ecotype activity (rRNA/rDNA) and relative abundance (rDNA%) for Ia (white circles), Ib (grey squares) and II (black triangles) and (b) relationship of rRNA to rDNA ratios between SAR11 ecotypes. Dashed line is 1:1. Correlation statistics are Ia and Ib (R=0.56; P<0.001); Ib and II (R=0.47; P<0.01); and Ia and II (R=0.38; P<0.05).
Discussion
Mixing controls surface bacterioplankton diversity maxima at broad phylogenetic levels
A striking result of our time-series analysis was the observation that levels of diversity measured by the Shannon Index, a metric that considers equitability as well as richness, showed repeatable minima occurring in the summer and maxima during winter months. These trends are remarkably similar when all OTUs were compared against only those within the SAR11 clade. Our findings are in agreement with the emerging paradigm of strong and predictable seasonality in total prokaryotic diversity (Gilbert et al., 2012; Ladau et al., 2013). However, the concordance between the dynamics of the entire community that contains a physiologically diverse assemblage of OTUs, to that of a narrower clade such as SAR11, suggests significant forcing by physical mechanisms which act similarly across broad phylogenetic levels.
We propose water column mixing to be the primary mechanism regulating seasonal augmentation of diversity in the NW Mediterranean. Periods of high and low bacterial and SAR11 ecotype diversity were repeatedly associated with deep and shallow mixed layers, respectively (Table 1). Furthermore, MLD was quantitatively the most important environmental parameter for predictive modelling of measured SAR11 ecotype sequence abundance. Presumably, stratification breakdown directly enhances diversity simply by mixing vertically segregated populations in a ‘community-merging’ event. Microbial populations frequently exhibit environmental niche partitioning as a function of depth (Field et al., 1997; West and Scanlan, 1999; DeLong et al., 2006; Carlson et al., 2009; Treusch et al., 2009) and convective overturning has been shown to be important for the temporal and vertical distributions of certain bacterial lineages (Morris et al., 2005; Treusch et al., 2009; Carlson et al., 2009; Eiler et al., 2009; Chow et al., 2013; Vergin et al., 2013b). In addition, we find seasonal maxima of SAR11 diversity is unrelated to the activity of these ecotypes estimated from their rRNA/rDNA ratios, further supporting the idea of translocation from biogeochemically distinct niches throughout the water column. Similar to findings in the English Channel (Gilbert et al., 2012) we observe a strong correlation between diversity and day-length (data not shown). Although a good-predictor of bacterial diversity, we suggest that day-length is a non-mechanistic correlate of seasonality. Changes in day-length integrate strongly seasonal features, including mixing.
Subsequent to the process of community merging, mixing the water column may lead to two other mechanistic processes; ‘ex-novo substrate merging’ and ‘de-novo substrate induction’, which act to consolidate high diversity systems. Substrate merging relies on the fact that the wide spectrum of substrates available for bacterial growth exhibits vertical structure (Carlson et al., 2004; Reinthaler et al., 2006). Mixing increases the range of substrates, and thus bacterial diversity, by rapidly expanding the number of available niches in the surface layer. Substrate induction may result from the production of new organic substrates by eukaryotic blooms stimulated by macronutrient resupply upon mixing. At our study site there is a strong association of enhanced chlorophyll features with deeper mixed layers. The potential range of de novo substrates produced from variable eukaryotic sources (Nelson et al., 2013; Sarmento et al., 2013) could also expand the number of bacterial niches (Landa et al., 2013) in the surface ocean. However, in the NW Mediterranean peaks in chlorophyll tend to succeed those of bacterial and SAR11 diversity, suggesting it has a second order effect. Water column mixing (and the combination of populations that inhabited different strata during summer months) is therefore the most parsimonious explanation to explain the origin of winter diversity maxima in the NW Mediterranean, and perhaps those observed in other temperate environments (for example, see Gilbert et al., 2012; Ladau et al., 2013; Vergin et al., 2013b). Ex-novo substrate merging and de novo substrate induction are suggested as secondary mechanisms related to physical mixing that act to consolidate high diversity states for some interval subsequent to the initial community-merging event.
Although it is intuitive that physical mixing of depth separated niche space enhances SAR11 ecotype diversity, it is more difficult to envisage how shoaling of the mixed layer could directly select against certain ecotypes. Stratification must influence SAR11 ecotype dynamics indirectly through competitive selection (for example, growth, substrate limitation, grazing, viral lysis). We thus propose a framework in which physical mixing is a common mechanism responsible for periodically ‘resetting’ bacterial ecosystems to a high diversity state. The subsequent progression towards low diversity communities commonly observed (Treusch et al., 2009; Gilbert et al., 2012; Chow et al., 2013) is driven by seasonal evolution of local environmental conditions, leading to the dominance of more oligotrophic and high light-adapted SAR11 ecotypes. These environmental conditions may vary between regions and affect bacterial sub-groups in different ways. However, the widely reported winter maxima in bacterial diversity (Gilbert et al., 2012; Ladau et al., 2013) suggest increases in diversity due to vertical mixing are a widespread phenomenon in temperate environments. The seasonal diversity patterns observed in the NW Mediterranean and other temperate (Gilbert et al., 2012; Ladau et al., 2013) and high latitude environments (Ghiglione et al., 2012) highlight the importance of adequate temporal resolution when attempting to synthesise global patterns of bacterial diversity.
Environmental selection of low diversity regimes
In the NW Mediterranean we propose inorganic nutrient limitation, specifically that of phosphate, as a plausible factor driving SAR11 towards a low diversity state. SAR11 Ia and Ib/II relative abundances were inversely correlated with nutrient concentrations. In the NW Mediterranean phosphate concentrations are very low (38±17 nM) and frequently close to analytical detection limits. In such extreme oligotrophic environments turnover is a more reliable metric of phosphate limitation. Low diversity SAR11 communities occur congruently with periods of very rapid (<5 h) phosphate turnover. In addition, SAR11 ecotypes were differentially correlated with Synechococcus abundance. Synechococcus are known to have high cell-specific uptake rates for PO43− (Casey et al., 2009; Michelou et al., 2011) and may enhance phosphate limitation, promoting the development of low diversity SAR11 communities dominated by group Ia. In support of our environmental data, genomic sequences point to the adaptation of SAR11 Ia to rapid [PO43−] turnover through enhanced expression of high-affinity phosphate transporters (Wilhelm et al., 2007; Sowell et al., 2008) or the ability to hydrolyse organic phosphorous substrates (Grote et al., 2012). Unfortunately sequence information from groups Ib and II is currently lacking so the hypothesis of phosphorus as a segregating factor for SAR11 ecotypes remains circumstantial.
The dominance of SAR11 Ia also corresponds to seasonal maxima in leucine incorporation rates (Figure 3b). Dissolved organic matter accumulates in highly stratified surface layers (Williams, 1995; Hansell and Carlson, 1998) including our study site (Laghdass et al., 2012). Low diversity communities dominated by SAR11 Ia thus appear capable of significant dissolved organic carbon utilisation at low nutrient concentrations, resulting in enhanced bacterial production relative to other times of year. Temperature effects and leucine uptake by co-occurring bacterial groups, including cyanobacteria, may also contribute to enhanced bacterial metabolism during summer.
Predictive model of SAR11 ecotype sequences
The available biogeochemical observations were used to calibrate a PLSR model that was capable of describing 70% of cross-validated variance (Q2) in the temporal dynamics of SAR11 ecotype groupings. Mixed layer depth emerged as the most important environmental control. Overall, regression coefficients between the measured biogeochemical parameters and tag sequence data were larger for the rRNA fraction, perhaps indicating tighter bottom-up control on active SAR11 ecotypes. SAR11 Ia dominates in shallow mixed layers of mid-late summer that are depleted in inorganic nutrients, have low chlorophyll biomass and picoplankton populations dominated by Synechococcus. Groups Ib and II are associated with deeper mixed layers enhanced in nutrients and phytoplankton biomass. It is presently unclear however if ecotypes Ib and II are specifically adapted to enhanced phytoplankton biomass and nutrients in the same way as other bacterial clades; for example, Roseobacter (Le Fort and Gasol, 2013). The persistence of these clades for several months may be a result of repeated ‘physical-seeding’ to the surface throughout the mixing season.
Activity of SAR11 ecotypes from rRNA/rDNA ratios
The ratio of rRNA and ribosomal RNA gene copies (rRNA/rDNA) is frequently used to estimate in-situ activity, or even growth rates, of specific bacterial OTUs (Campbell et al., 2011; Campbell and Kirchman, 2013; Hunt et al., 2013). However, a recent report has questioned the reliability of rRNA as an indicator of metabolic state (Blazewics et al., 2013). To validate the utility of rRNA/rDNA in describing in-situ variation of SAR11 ecotype activity we compared tag sequence data to direct measurements of leucine incorporation (Laghdass et al., 2012), which is a commonly used measure of heterotrophic production by bacteria (Kirchman et al., 1985). The fraction of SAR11 incorporating leucine was positively correlated with SAR11 rRNA/rDNA ratio. Although limited in number, our data set covers the full range of rRNA/rDNA ratios (Campbell et al., 2009, 2011) and leucine incorporation rates (for example, Malmstrom et al., 2007) typically reported for SAR11. To the best of our knowledge we have made the first attempt to correlate rRNA/rDNA as an in-situ activity metric against a direct measure of bacterial metabolism at an identical, albeit broad, phylogenetic resolution. It is difficult to speculate if the relationship observed for SAR11 would hold for other bacterial lineages or at increasing phylogenetic resolution. From the limited observations we make a first-order assumption that rRNA/rDNA and leucine incorporation exhibit similar covariance at ecotype level and therefore apply this ratio to investigate patterns in SAR11 ecotype activity.
The rRNA/rDNA ratios of SAR11 ecotypes did not display any coherent seasonal patterns or relationships with the measured biogeochemical variables (Supplementary Figure 2; Table 1). Furthermore, ecotype abundance and activity were uncorrelated (Figure 6a). It might be expected that correlations between ecotype activity and abundance are characterised by an undefined time lag. However, applying a one-month time lag [+1 sampling event] (for example, Fuhrman et al., 2006) to our data set failed to improve activity-abundance correlations. Decoupling of activity and abundance has been observed elsewhere (Campbell and Kirchman, 2013). The temporal resolution afforded by a monthly time-series may be too coarse to capture the transient nature of abundance–activity relationships in SAR11 ecotypes.
Alternatively, it is possible that despite differences in 16S RNA gene phylogeny (Vergin et al., 2013a) and gene content (Grote et al., 2012), SAR11 ecotypes share enough metabolic potential and overlapping niche space to maintain simultaneously active populations. In support of this SAR11 ecotype activities are statistically indistinguishable from each other in low and high diversity settings, regardless of the strong biogeochemical gradients that exist. Positive covariance in SAR11 ecotype activities across the 7-year data set (Figure 6b) also suggest that activity of different ecotypes may respond similarly to changes in environmental conditions. However, the clear seasonal partitioning of SAR11 ecotype abundances are not systematically related to specific ecotype activity, as presently determined from rRNA/rDNA ratios. Although we provide preliminary evidence that potential metabolic activity (rRNA abundance) is linked to realised protein synthesis (leucine incorporation) in SAR11, the use of rRNA/rDNA ratios as a reliable activity measure remains ambiguous (Blazewics et al., 2013).
Through our analysis we demonstrate how physical oceanographic processes and biogeochemical forcing factors combine to explain the well-documented seasonal cycles in bacterial diversity. Temporal instability in environmental conditions is the main factor driving patterns of bacterial diversity in the NW Mediterranean. We suggest that adequate spatial and temporal global coverage would reveal bacterial diversity peaks in winter at temperate to high latitudes, rather than seasonally stable low latitude environments. Although our analysis specifically focuses on SAR11 dynamics we expect the same mechanisms to be relevant for other bacterial clades. In the NW Mediterranean phosphate limitation appears to govern seasonal transitions to low-diversity SAR11 communities. Phosphate may be of similar importance in other oligotrophic environments that are characterised by seasonal dominance of SAR11 ecotype Ia. PLSR modelling demonstrated strong and predictable temporal patterns in SAR11 ecotype abundance that were not reflected in seasonal dynamics of metabolic activity. Future studies targeting the fine-scale phylogenetic structure of SAR11 communities, at increased temporal resolution, may help to further define the complex patterns observed between SAR11 ecotype abundance and activity.
Accession codes
References
Alonso-Sáez LV, Balagué E, Sà O, Sánchez JM, González J, Pinhassi R et al. (2007). Seasonality in bacterial diversity in NW Mediterranean coastal waters: assessment through clone libraries, fingerprinting and fluorescence in situ hybridization. FEMS Microbiol Ecol 60: 98–112.
Alonso-Sáez LV, Gasol JP . (2007). Seasonal variations in the contributions of different bacterial groups to the uptake of low-molecular-weight compounds in Northwestern Mediterranean Coastal Waters. Appl Environ Microbiol 73: 3528–3535.
Beman JM, Steele JA, Furhman JA . (2011). Co-occurrence patterns for abundant marine archaeal and bacterial lineages in the deep chlorophyll maximum of coastal California. ISME J 5: 1077–1085.
Benjamini Y, Hochberg Y . (1995). Controlling the false discovery Rate: A practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300.
Blanquer A, Uriz MJ, Galand PE . (2013). Removing environmental sources of variation to gain insight on symbionts vs. transient microbes in high and low microbial abundance sponges. Environ Microbiol 15: 3008–3019.
Blazewics SJ, Barnard RL, Daly RA, Firestone MK . (2013). Evaluating rRNA as an indicator of microbial activity in environmental communities: limitations and uses. ISME J 7: 2061–2068.
Brettar I, Christen R, HöFle MG . (2011). Analysis of bacterial core communities in the central Baltic by comparative RNA–DNA-based fingerprinting provides links to structure–function relationships. ISME J 6: 195–212.
Brown MV, Fuhrman JA . (2005). Marine bacterial microdiversity as revealed by internal transcribed spacer analysis. Aquat Microb Ecol 41: 15–23.
Brown MV, Lauro FM, DeMaere MZ, Muir L, Wilkins D, Thomas T et al. (2012). Global Biogeography of SAR11 marine bacteria. Mol Syst Biol 8: 595.
Campbell BJ, Kirchman DL . (2013). Bacterial diversity, community structure and potential growth rates along an estuarine salinity gradient. ISME J 7: 210–220.
Campbell BJ, Yu L, Heidelberg JF, Kirchman DL . (2011). Activity of abundant and rare bacteria in a coastal ocean. Proc Natl Acad Sci USA 108: 12776–12781.
Campbell BJ, Yu L, Straza TRA, Kirchman DL . (2009). Temporal changes in bacterial rRNA and rRNA genes in Delaware (USA) coastal waters. Aquat Microb Ecol 57: 123–135.
Carlson CA, Giovannoni SJ, Hansell DA, Goldberg SJ, Parsons R, Vergin K . (2004). Interactions among dissolved organic carbon, microbial processes, and community structure in the mesopelagic zone of the northwestern Sargasso Sea. Limnol Oceanogr 49: 1073–1083.
Carlson CA, Morris R, Parsons R, Treusch AH, Giovannoni SJ, Vergin K . (2009). Seasonal dynamics of SAR11 populations in the euphotic and mesopelagic zones of the northwestern Sargasso Sea. ISME J 3: 283–295.
Casey JR, Lomas MW, Michelou VK, Dyhrman ST, Orchard ED, Ammerman JW et al. (2009). Phytoplankton taxon-specific orthophosphate (Pi) and ATP utilization in the western subtropical North Atlantic. Aquat Microb Ecol 58: 31–44.
Chow CET, Sachdeva R, Cram JA, Steele JA, Needham DM, Patel A et al. (2013). Temporal variability and coherence of euphotic zone bacterial communities over a decade in the Southern California Bight. ISME J 7: 2259–2273.
DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU et al. (2006). Community genomics among stratified microbial assemblages in the ocean’s interior. Science 311: 496–503.
DeLong EF, Wickham GS, Pace NR . (1989). Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 243: 1360–1363.
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072.
Edgar RC . (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797.
Edgar RC . (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461.
Eiler A, Hayakawa DH, Church MJ, Karl DM, Rappe MS . (2009). Dynamics of the SAR11 bacterioplankton lineage in relation to environmental conditions in the oligotrophic North Pacific subtropical gyre. Environ Microbiol 11: 2291–2300.
Fegatella F, Lim J, Kjelleberg S, Cavicchioli R . (1998). Implications of rRNA operon copy number and ribosome content in the marine oligotrophic ultramicrobacterium Sphingomonas sp. strain RB2256. Appl Environ Microbiol 64: 4433–4438.
Felsenstein J . (2008) PHYLIP (Phylogeny Inference Package) version 3.68. Distributed by the author. Department of Genome Sciences. University of Washington: Seattle.
Field KG, Gordon D, Wright T, Rappé M, Urback E, Vergin K et al. (1997). Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl Environ Micro 63: 63–70.
Fuhrman JA, Hewson I, Schwalbach MS, Steele JA, Brown MV, Naeem S . (2006). Annually reoccurring bacterial communities are predictable from ocean conditions. Proc Natl Acad Sci USA 103: 13104–13109.
Fuhrman JA . (2009). Microbial community structure and its functional implications. Nature 459: 193–199.
Galdos E, Rusch A, Ilardo M . (2011). Ribosomal tag pyrosequencing of DNA and RNA from benthic coral reef microbiota: community spatial structure, rare members and nitrogen-cycling guilds. Environ Microbiol 13: 1138–11522.
Garcia-Martinez J, Rodriguez-Valera F . (2000). Microdiversity of uncultured marine prokaryotes: the SAR11 cluster and the marine archaea of group I. Mol Ecol 9: 935–948.
Gentile G, Giuliano L, D’Auria G, Smedile F, Azzaro M, De Domenico M et al. (2006). Study of bacterial communities in Antarctic coastal waters by a combination of 16S rRNA and 16S rDNA sequencing. Environ Microbiol 8: 2150–2161.
Ghiglione JF, Galand PE, Pommier T, Pedrós-Alió C, Maas EW, Bakker K et al. (2012). Pole to pole biogeography of surface and deep marine bacterial communities. Proc Natl Acad Sci USA 109: 17633–17638.
Gilbert JA, Field D, Swift P, Newbold L, Oliver A, Smyth T et al. (2009). The seasonal structure of microbial communities in the Western English Channel. Environ Microbiol 11: 3132–3139.
Gilbert JA, Steele JA, Caporaso JG, Steinbruck L, Reeder J, Temperton B et al. (2012). Defining seasonal marine microbial community dynamics. ISME J 6: 298–308.
Giovannoni SJ, Bibbs L, Cho JC, Stapels MD, Desiderio R, Vergin KL et al. (2005b). Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature 438: 82–85.
Giovannoni SJ, Rappé MS . (2000). Evolution, diversity and molecular ecology of marine prokaryotes. In: Kirchman DL, (ed) Microbial Ecology of the Oceans. Wiley and Sons, New York, NY pp 47–84.
Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D et al. (2005a). Genome streamlining in a cosmopolitan oceanic bacterium. Science 309: 1242–1245.
Giovannoni SJ, Vergin KL . (2012). Seasonality in ocean microbial communities. Science 335: 671–676.
Grote J, Thrash JC, Huggett MJ, Landry ZC, Carini P, Giovammoni SJ et al. (2012). Streamlining and core genome conservation among highly divergent members of the SAR11 Clade. Mbio 3: pii e00252–12.
Hammer Ø, Harper DAT, Ryan PD . (2001). PAST: paleontological statistics software package for education and data analysis. Palaeo Electronica 4.
Hansell DA, Carlson CA . (1998). Net community production of dissolved organic carbon. Global Biogeochem Cycles 12: 443–453.
Hugoni M, Taib N, Debroas D, Domaizon I, Dufournel IJ, Bronner G et al. (2013). Structure of the rare archaeal biosphere and seasonal dynamics of active ecotypes in surface coastal waters. Proc Natl Acad Sci USA 110: 6004–6009.
Hunt DE, Lin Y, Church MJ, Karl DM, Tringe SG, Izzo LK et al. (2013). Relationship between abundance and specific activity of bacterioplankton in open ocean surface waters. Appl Environ Microbiol 79: 177–184.
Huse SM, Welch DM, Morrison HG, Sogin ML . (2010). Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol 12: 1889–1898.
Kerkhof L, Kemp P . (1999). Small ribosomal RNA content in marine Proteobacteria during non-steady-state growth. FEMS Microbiol Ecol 30: 253–260.
Kirchman D, K’nees E, Hodson R . (1985). Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural aquatic systems. Appl Environ Microbiol 49: 599–607.
Kirchman D . (1993). Leucine incorporation as a measure of biomass production by heterotrophic bacteria. In: Kemp PF, Sherr BF, Sherr EB, Cole JJ, (eds) Handbook of Methods in Aquatic Microbial Ecology. Lewis: Boca Raton, pp 509–512.
Koeppel A, Perry EB, Sikorski J, Krizanc D, Warner A, Ward D et al. (2008). Identifying the fundamental units of bacterial diversity: a paradigm shift to incorporate ecology into bacterial systematics. Proc Natl Acad Sci USA 105: 2504–2509.
Kunin V, Engelbrektson A, Ochman H, Hugenholtz P . (2010). Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol 12: 118–123.
Kunin V, Hugenholtz B . (2010). PyroTagger: a fast, accurate pipeline for analysis of rRNA amplicon pyrosequence data. Open J 1: 1–8.
Ladau J, Sharpton TJ, Finucane MM, Jospin G, Kembel SW, O’Dwyer J et al. (2013). Global marine bacterial diversity peaks at high latitudes in winter. ISME J 7: 1669–1677.
Laghdass M, Catala P, Caparros J, Oriol L, Lebaron P, Obernosterer I . (2012). High contribution of SAR11 to microbial activity in the North West Mediterranean Sea. Microb Ecol 63: 324–333.
Laghdass M, West NJ, Batailler N, Caparros J, Catala P, Lantoine F et al. (2010). Impact of lower salinity waters on bacterial heterotrophic production and community structure in the offshore NW Mediterranean Sea. Environ Microbiol Reports 2: 761–769.
Lami R, Ghiglione JF, Desdevises Y, West NJ, Lebaron P . (2009). Annual patterns of presence and activity of marine bacteria monitored by 16S rDNA–16S rRNA fingerprints in the coastal NW Mediterranean Sea. Aquat Microb Ecol 54: 199–210.
Landa M, Cottrell M, Kirchman D, Blain S, Obernosterer I . (2013). Changes in bacterial diversity in response to dissolved organic matter supply in a continuous culture experiment. Aquat Microb Ecol 69: 157–168.
Lefort T, Gasol JM . (2013). Global-scale distributions of marine surface bacterioplankton groups along gradients of salinity, temperature, and chlorophyll: a meta-analysis of fluorescence in situ hybridization studies. Aquat Microb Ecol 70: 111–130.
Malmstrom RR, Cottrell MT, Elifantz H, Kirchman DL . (2005). Biomass production and assimilation of dissolved organic matter by SAR11 bacteria in the northwest Atlantic Ocean. Appl Environ Microbiol 71: 2979–2986.
Malmstrom RR, Kiene RP, Cottrell MT, Kirchman DL . (2004). Contribution of SAR11 bacteria to dissolved dimethylsulfoniopropionte and amino acid uptake in the North Atlantic Ocean. Appl Environ Microbiol 70: 4129–4135.
Malmstrom RR, Straza TRA, Cottrell MT, Kirchman DL . (2007). Diversity, abundance and biomass production of bacterial groups in the western Arctic Ocean. Aquat Microb Ecol 47: 45–55.
Michelou VK, Lomas MW, Kirchman DL . (2011). Phosphate and adenosine-5′triphosphate uptake by cyanobacteria and heterotrophic bacteria in the Sargasso Sea. Limnol Oceanogr 56: 323–332.
Moeseneder MM, Winter C, Herndl GJ . (2001). Horizontal and vertical complexity of attached and free-living bacteria of the eastern Mediterranean Sea, determined by 16S rDNA and 16S rRNA fingerprints. Limnol Oceanogr 46: 95–107.
Morris RM, Rappé MS, Connon SA, Vergin KL, Siebold WA, Carlson CA et al. (2002). SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420: 806–810.
Morris RM, Vergin KL, Cho JC, Rappe MS, Carlson CA, Giovannoni SJ . (2005). Temporal and spatial response of bacterioplankton lineages to annual convective overturn at the Bermuda Atlantic Time-series Study site. Limnol Oceanogr 50: 1687–1696.
Nelson CE, Goldberg SJ, Wegley LW, Haas AF, Smith JE, Rohwer F et al. (2013). Coral and macroalgal exudates vary in neutral sugar composition and differentially enrich reef bacterioplankton lineages. ISME J 7: 962–979.
Reinthaler T, van Aken H, Veth C, Arístegui J, Robinson C, Williams PJ le B et al. (2006). Prokaryotic respiration and production in the meso- and bathypelagic realm of the eastern and western North Atlantic basin. Limnol Oceanogr 51: 1262–1273.
Rusch D, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S et al. (2007). The Sorcerer II Global Ocean sampling expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol 5: 398–431.
Sarmento H, Romero-Castillo C, Lindh H, Pinhassi J, Sala MM, Gasol JM et al. (2013). Phytoplankton species-specific release of dissolved free amino acids and their selective consumption by bacteria. Limnol Oceanogr 58: 1123–1135.
Schauer M, Balagué V, Pedrós-Alió C, Massana R . (2003). Seasonal changes in the taxonomic composition of bacterioplankton in a coastal oligotrophic system. Aquat Microb Ecol 31: 163–174.
Smith DC, Azam F . (1992). A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine. Mar Microb Food Webs 6: 107–114.
Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR et al. (2006). Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. Proc Natl Acad Sci USA 103: 12115–12120.
Solorzano L . (1969). Determination of ammonia in natural waters by the phenol-hypochlorite method. Limnol Oceanogr 14: 799–801.
Sowell S, Wilhelm L, Norbeck A, Lipton M, Nicora C, Barofsky D et al. (2008). Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME J 3: 93–105.
Stingl U, Tripp HJ, Giovannoni SJ . (2007). Improvements of high throughput culturing yielded novel SAR11 strains and other abundant marine bacteria from the Oregon coast and the Bermuda Atlantic Time Series study site. ISME J 1: 361–371.
Strickland JD, Parsons TR . (1997) A Practical Handbook of Seawater Analysis 2nd edn Bulletin of the Fisheries Research Board of Canada.
Suzuki MT, Béjà O, Taylor LT, DeLong EF . (2001). Phylogenetic analysis of ribosomal RNA operons from uncultivated coastal marine bacterioplankton. Environ Microbiol 3: 323–331.
Treguer P, Le Corre P . (1975) Manuel d’analyse des Sels Nutritifs Dans L'eau de mer Utilisation de l'AutoAnalyzer 2 Technicon 2nd edn. 'Univ Bretagne occidentale, Brest.
Treusch AH, Vergin KL, Finlay LA, Donatz MG, Burton RM, Carlson C et al. (2009). Seasonality and vertical structure of microbial communities in an ocean gyre. ISME J 3: 1148–1163.
Troussellier M, Schäfer H, Batailler N, Bernard L, Courties C, Lebaron P et al. (2002). Bacterial activity and genetic richness along an estuarine gradient (Rhone River plume, France). Aquat Microb Ecol 28: 13–24.
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA et al. (2004). Environmental genome shotgun sequencing of the Sargasso Sea. Science 304: 66–74.
Vergin KL, Beszteri B, Monier A, Thrash JC, Temperton B, Treusch AH . (2013a). High-resolution SAR11 ecotype dynamics at the Bermuda Atlantic Time-Series Study Site by phylogenetic placement of pyrosequences. ISME J 7: 1322–1332.
Vergin KL, Done B, Carlson CA, Giovannoni SJ . (2013b). Spatiotemporal distributions of rare bacterioplankton populations indicated adaptive strategies in the oligotrophic ocean. Aquat Microb Ecol 71: 1–13.
West NJ, Obernosterer I, Zemb O, Lebaron P . (2008). Major differences of bacterial diversity and activity inside and outside of a natural iron-fertilized phytoplankton bloom in the Southern Ocean. Environ Microbio 10: 738–756.
West NJ, Scanlan DJ . (1999). Niche-partitioning of Prochlorococcus populations in a stratified water column in the Eastern North Atlantic Ocean. Appl Environ Microbiol 65: 2585–2591.
Wilhelm L, Tripp H, Givan S, Smith D, Giovannoni S . (2007). Natural variation in SAR 11 marine bacterioplankton genomes inferred from metagenomic data. Biol Direct 2: 27.
Williams PJ . (1995). Evidence for the seasonal accumulation of carbon-rich dissolved organic material, its scale in comparison with changes in particulate material and the consequential effect on net C/N assimilation ratios. Mar Chem 51: 17–29.
Wold S, Sjöström M, Eriksson L . (2001). PLS regression: a basic tool of chemometrics. Chemom Intell Laboratory Syst 58: 109–130.
Wright RT, Hobbie JE . (1966). Use of glucose and acetate by bacteria and algae in aqautic ecosystems. Ecology 47: 447–464.
Yentsch CS, Menzel DW . (1963). Method for the determination of phytoplankton chlorophyll and phaeophytin by fluorescence. Deep-Sea Res 10: 221–231.
Zubkov MV, Mary I, Woodward EMS, Warwick PE, Fuchs BM, Scanlan DJ et al. (2007). Microbial control of phosphate in the nutrient-depleted North Atlantic subtropical gyre. Environ Microbiol 9: 2079–2089.
Acknowledgements
We thank Eric Maria and Louise Oriol for assisting with the collection and analysis of samples for the physical and chemical parameters over the time-series. The work was jointly supported by a LEFE-CYBER grant awarded to Ian Salter and LOMIC micro-project grant awarded to Ian Salter. We acknowledge Platform BIO2MAR for the provision of molecular biology equipment.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Salter, I., Galand, P., Fagervold, S. et al. Seasonal dynamics of active SAR11 ecotypes in the oligotrophic Northwest Mediterranean Sea. ISME J 9, 347–360 (2015). https://doi.org/10.1038/ismej.2014.129
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2014.129
This article is cited by
-
Dynamics of actively dividing prokaryotes in the western Mediterranean Sea
Scientific Reports (2022)
-
Influence of short and long term processes on SAR11 communities in open ocean and coastal systems
ISME Communications (2022)
-
Spatial and diel variations of the prokaryotic community in the Phaeocystis globosa blooms area of Beibu Gulf, China
Acta Oceanologica Sinica (2022)
-
Temporal and spatial dynamics of Bacteria, Archaea and protists in equatorial coastal waters
Scientific Reports (2019)
-
Distribution Patterns of Microbial Community Structure Along a 7000-Mile Latitudinal Transect from the Mediterranean Sea Across the Atlantic Ocean to the Brazilian Coastal Sea
Microbial Ecology (2018)