Abstract
Dysbiotic oral bacterial communities have a critical role in the etiology and progression of periodontal diseases. The goal of this study was to investigate the extent to which smoking increases risk for disease by influencing the composition of the subgingival microbiome in states of clinical health. Subgingival plaque samples were collected from 200 systemically and periodontally healthy smokers and nonsmokers. 16S pyrotag sequencing was preformed generating 1 623 713 classifiable sequences, which were compared with a curated version of the Greengenes database using the quantitative insights into microbial ecology pipeline. The subgingival microbial profiles of smokers and never-smokers were different at all taxonomic levels, and principal coordinate analysis revealed distinct clustering of the microbial communities based on smoking status. Smokers demonstrated a highly diverse, pathogen-rich, commensal-poor, anaerobic microbiome that is more closely aligned with a disease-associated community in clinically healthy individuals, suggesting that it creates an at-risk-for-harm environment that is primed for a future ecological catastrophe.
Similar content being viewed by others
Introduction
One of the first ecosystems to come into contact with tobacco smoke, and to be affected by it, is the oral microbiome. Smokers present a high-at-risk cohort for periodontitis (Tomar and Asma, 2000), a microbially driven oral disease that affects two-thirds of adult humans (Eke et al., 2012). We have previously demonstrated that smoking is associated with pathogen enrichment in periodontal disease (Shchipkova et al., 2010) and that smokers acquire these pathogens within 24 h of biofilm formation (Kumar et al., 2011b). The goal of the present study, therefore, was to investigate whether smoking has a role in the etiopathogenesis of periodontitis by creating an at-risk-for-harm subgingival microbiome in states of clinical health.
The study was approved by the Office of Research at The Ohio State University (2008H0122) and the NHS National Research Ethics Service Northeast (09/H0904/46). Two hundred periodontally healthy individuals between 21 and 40 years of age were recruited. Subjects who reported diabetes, HIV, pregnancy, immunosuppressants, bisphosphonates, steroids, current orthodontic therapy, antibiotics or professional dental cleaning within 3 months, and those requiring pretreatment antibiotic coverage were excluded. Subjects completed a demographic and tobacco exposure questionnaire and were examined by calibrated periodontists. Subjects had least 20 natural non-carious teeth, ⩽3 mm probing pocket depths at all sites, plaque index of ⩽0.9 (Loe et al., 1965) and gingival index of ⩽1.2 (Loe and Silness, 1963). Both groups were frequency-matched for age, gender, race/ethnicity, education and socioeconomic status (Supplementary Table 1).
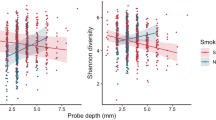
Subgingival plaque was collected by inserting endodontic paper points into the interproximal gingival sulci of 10 randomly selected teeth and pooled. Bacterial DNA was isolated (Kumar et al., 2012) and 16S pyrotag sequencing was performed (Dowd et al., 2008) in a commercial facility (Research and Testing Laboratories, Lubbock, TX, USA). For each sample, V1–V3 and V7–V9 regions of the 16S gene were sequenced and combined to create a composite data set (Kumar et al., 2011a) and compared with a locally hosted version of Greengenes (DeSantis et al., 2006) using the quantitative insights into microbial ecology pipeline (Caporaso et al., 2010) and locally developed complimentary tool set qiime-tools (http://github.com/smdabdoub/qiime-tools) for phylogenetic analysis. The sequences are deposited in the Sequence Read Archive database of NCBI (accession number SRS590909). In all, 1 581 383 denoised and chimera-depleted sequences were classified with 97% similarity to 572 species-level operational taxonomic units (s-OTUs); with 155±27 s-OTUs in each individual. Smokers demonstrated a remarkable similarity in microbial composition (Figure 1a), as evidenced by significant lineage-based clustering using principal coordinate analysis (PCoA) (P<0.001, analysis of similarity (ANOSIM)). We have previously shown that ethnic origin may be a significant determinant of subgingival microbial composition (Mason et al., 2013). However, when a subset of 50 Caucasians (Figure 1b) and 34 African Americans (Figure 1c) was analyzed by PCoA, both ethnic groups clustered by smoking status. In contrast, when a subset of 17 African-American and 25 Caucasian smokers (Figure 1d) was compared by PCoA no clustering was observed, indicating that smoking supersedes the influence of genotypic factors such as ethnicity on the microbiome.
PCoA plots of the UniFrac distance for all samples by smoking status. (a) All 200 samples, a subset of (b) 50 Caucasians and (c) 34 African Americans are shown. All samples demonstrated significant clustering based on tobacco exposure, irrespective of ethnic background (P<0.05, analysis of similarity (ANOSIM) on unweighted UniFrac distances). (d) A subset of 17 African-American and 25 Caucasian smokers that does not demonstrate significant clustering based on tobacco exposure.
A principle trait of a health-compatible community is niche saturation, an ecological phenomenon where a few species dominate the community, and resist colonization by pathogenic organisms (Brockhurst et al., 2007), thus maintaining mucosal health (Abt and Pamer, 2014). Increase in species diversity has been observed in periodontal disease (Loe et al., 1965; Listgarten, 1976; Loesche and Syed, 1978), and high diversity drives further speciation (Emerson and Kolm, 2005), leading to perpetuation of disease. Shannon diversity index incorporates both the number of s-OTUs (richness) and relative abundance of each s-OTU (evenness) into a single value. Whereas a diversity index of zero represents a mono-species community, a higher value may result either from the presence of several species or from equitable distribution of a few species. Smokers demonstrated a significantly higher Shannon diversity index (4.85 versus 4.35, P=0.01, Welch analysis of variance), and lower variance (as measured by the standard deviations on the diversity index) than nonsmokers (P=0.0002, Levene’s test), indicating that each smoker harbored more numbers of s-OTUs than a nonsmoker; however, as a group, current smokers demonstrate less heterogeneity in the composition of the subgingival microbiome. Thus, with respect to diversity, the microbiome of a clinically healthy smoker demonstrated the characteristics of a community associated with disease. Thus, smoking appears to create a microenvironment that selects for a large and specific group of micro-organisms; and by doing so, may decrease the niche saturation abilities of this community. This loss of microbial protection may be one mechanism by which smoking increases the risk for disease.
Smokers demonstrated higher abundances of anaerobes and lower levels of aerobes when compared with nonsmokers (P=0.02, t-test) (Figure 2a, innermost circle). Also, 67 of the 128 genera were significantly different in abundances between groups, as were 172 s-OTUs. The subgingival microbiome of smokers was enriched for periodontal and systemic pathogens Fusobacterium nucleatum, F. naviforme, Filifactor alocis, Dialister microaerophilus, Desulfobulbus sp. clone R004, Megasphaera sueciensis, M. geminatus, M. elsdenii, M. micronuciformis, Acinetobacter johnsonii, A. guillouiae, A. schindleri, A. baumannii, A. haemolyticus, Pseudomonas pseudoalcaligenes and Pseudoramibacter alactolyticus (Figure 2a outermost circle, Figure 2b, and Supplementary Table 2), with significantly lower levels of several health-compatible commensals, for example, Streptococcus sanguinis, S. parasanguinis, S. oralis, Granulicatella elegans, G. adiacens, Actinomyces viscosus, A. israelii, A. dentalis, Neisseria subflava and Hemophilus parainfluenzae (Figure 2a outermost circle, Figure 2c and Supplementary Table 2).
(a) Phylogenetic tree of the 572 s-OTUs. It was generated using the interactive tree of life web application (http://itol.embl.de/). The outermost circle indicates the normalized mean relative abundances of the s-OTUs in nonsmokers (green) and smokers (red). Overall abundances for each OTU are indicated by the black-to-gray circular gradient (middle circle). The Gram staining and oxygen requirement characteristics are represented by the innermost circle with Gram-negative aerobes (red), Gram-negative anaerobes (magenta), Gram-positive aerobes (blue), Gram-positive anaerobes (purple) and unknown characteristics (gray). (b, c) The relative abundance of selected species in current smokers and nonsmokers. PCoA plots of the UniFrac distances are shown, with the size of each bubble representing the abundance of the species in that sample.
F. nucleatum (Figure 2b) acts as a ‘bridging species’, enabling co-adhesion between early (commensal) and late (pathogenic) colonizers. It has recently been implicated in the etiology of colorectal cancer (Rubinstein et al., 2013), another disease for which smokers present a high-at-risk cohort (Giovannucci, 2001). The high abundance of this species in healthy smokers along with several pathogens suggests yet another mechanism by which smoking creates pathogen-rich oral biofilms. The high levels of this species in smokers may also suggest a mechanism by which smoking increases the risk for colorectal cancer, and warrants further investigation.
Another interesting observation was the high levels of F. alocis (Figure 2b) in periodontally healthy smokers. Earlier investigations by our group have identified F. alocis as a putative periodontal pathogen (Kumar et al., 2005) and several lines of evidence have emerged since, supporting this hypothesis (Griffen et al., 2012, Shaddox et al., 2012). F. alocis is an obligate anaerobe whose growth in vitro has been shown to be stimulated by oxidative stress (Aruni et al., 2011), which may explain the high levels observed in the present study.
S. mutans and Lactobacillus spp. (Figure 2a and Supplementary Table 2) are usually abundant in supragingival plaque of caries-active individuals (Gross et al., 2010). We observed a significant elevation of S. mutans and Lactobacillus salivarius in the subgingival community of smokers. L. salivarius is particularly interesting as it was present only in smokers. Various studies have demonstrated an increased risk of caries in smokers (Benedetti et al., 2013). The elevation of these cariogenic species could explain the high caries-risk in smokers, and requires further investigation.
In summary, clinically healthy smokers possess a highly diverse, pathogen-rich, commensal-poor, anaerobic microbiome that is more closely aligned with a disease-associated community. This work suggests that smoking has a role in creating an at-risk-for-harm microbiome, thus priming the oral environment for a downstream ‘ecological catastrophe’ (Marsh, 2003).
Accession codes
References
Abt MC, Pamer EG . (2014). Commensal bacteria mediated defenses against pathogens. Curr Opin Immunol 29C: 16–22.
Aruni AW, Roy F, Fletcher HM . (2011). Filifactor alocis has virulence attributes that can enhance its persistence under oxidative stress conditions and mediate invasion of epithelial cells by porphyromonas gingivalis. Infect Immun 79: 3872–3886.
Benedetti G, Campus G, Strohmenger L, Lingstrom P . (2013). Tobacco and dental caries: a systematic review. Acta Odontol Scand 71: 363–371.
Brockhurst MA, Colegrave N, Hodgson DJ, Buckling A . (2007). Niche occupation limits adaptive radiation in experimental microcosms. PLoS One 2: e193.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336.
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072.
Dowd SE, Sun Y, Wolcott RD, Domingo A, Carroll JA . (2008). Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) for microbiome studies: bacterial diversity in the ileum of newly weaned Salmonella-infected pigs. Foodborne Pathog Dis 5: 459–472.
Eke PI, Dye BA, Wei L, Thornton-Evans GO, Genco RJ, CDC Periodontal Disease Surveillance workgroup. (2012). Prevalence of periodontitis in adults in the United States: 2009 and 2010. J Dent Res 91: 914–920.
Emerson BC, Kolm N . (2005). Species diversity can drive speciation. Nature 434: 1015–1017.
Giovannucci E . (2001). An updated review of the epidemiological evidence that cigarette smoking increases risk of colorectal cancer. Cancer Epidemiol Biomarkers Prev 10: 725–731.
Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK et al. (2012). Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J 6: 1176–1185.
Gross EL, Leys EJ, Gasparovich SR, Firestone ND, Schwartzbaum JA, Janies DA et al. (2010). Bacterial 16S sequence analysis of severe caries in young permanent teeth. J Clin Microbiol 48: 4121–4128.
Kumar PS, Brooker MR, Dowd SE, Camerlengo T . (2011a). Target region selection is a critical determinant of community fingerprints generated by 16S pyrosequencing. PLoS One 6: e20956.
Kumar PS, Matthews CR, Joshi V, de Jager M, Aspiras M . (2011b). Tobacco smoking affects bacterial acquisition and colonization in oral biofilms. Infect Immun 79: 4730–4738.
Kumar PS, Mason MR, Brooker MR, O'Brien K . (2012). Pyrosequencing reveals unique microbial signatures associated with healthy and failing dental implants. J Clin Periodontol 39: 425–433.
Kumar PS, Griffen AL, Moeschberger ML, Leys EJ . (2005). Identification of candidate periodontal pathogens and benficial species using quantitative 16S clonal analysis. J Clin Microbiol 43: 3944–3955.
Listgarten MA . (1976). Structure of the microbial flora associated with periodontal health and disease in man. A light and electron microscopic study. J Periodontol 47: 1–18.
Loe H, Silness J . (1963). Periodontal disease in pregnancy. I. Prevalence and severity. Acta Odontol Scand 21: 533–551.
Loe H, Theilade E, Jensen SB . (1965). Experimental gingivitis in man. J Periodontol 36: 177–187.
Loesche WJ, Syed SA . (1978). Bacteriology of human experimental gingivitis: effect of plaque and gingivitis score. Infect Immun 21: 830–839.
Marsh PD . (2003). Are dental diseases examples of ecological catastrophes? Microbiology 149: 279–294.
Mason MR, Nagaraja HN, Camerlengo T, Joshi V, Kumar PS . (2013). Deep sequencing identifies ethnicity-specific bacterial signatures in the oral microbiome. PLoS One 8: e77287.
Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW . (2013). Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 14: 195–206.
Shaddox LM, Huang H, Lin T, Hou W, Harrison PL, Aukhil I et al. (2012). Microbiological characterization in children with aggressive periodontitis. J Dent Res 91: 927–933.
Shchipkova AY, Nagaraja HN, Kumar PS . (2010). Subgingival microbial profiles of smokers with periodontitis. J Dent Res 89: 1247–1253.
Tomar SL, Asma S . (2000). Smoking-attributable periodontitis in the United States: findings from NHANES III. National Health and Nutrition Examination Survey. J Periodontol 71: 743–751.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Mason, M., Preshaw, P., Nagaraja, H. et al. The subgingival microbiome of clinically healthy current and never smokers. ISME J 9, 268–272 (2015). https://doi.org/10.1038/ismej.2014.114
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2014.114
This article is cited by
-
Salivary microbiomes: a potent evidence in forensic investigations
Forensic Science, Medicine and Pathology (2024)
-
The mediating roles of the oral microbiome in saliva and subgingival sites between e-cigarette smoking and gingival inflammation
BMC Microbiology (2023)
-
Gut microbiome-based machine learning for diagnostic prediction of liver fibrosis and cirrhosis: a systematic review and meta-analysis
BMC Medical Informatics and Decision Making (2023)
-
Smoking and salivary microbiota: a cross-sectional analysis of an Italian alpine population
Scientific Reports (2023)
-
Smoking-induced subgingival dysbiosis precedes clinical signs of periodontal disease
Scientific Reports (2023)