Abstract
Novel lineages of the phylum Thaumarchaeota are endemic to thermal habitats, and may exhibit physiological capabilities that are not yet observed in members of this phylum. The primary goals of this study were to conduct detailed phylogenetic and functional analyses of metagenome sequence assemblies of two different thaumarchaeal populations found in high-temperature (65–72 °C), acidic (pH∼3) iron oxide and sulfur sediment environments of Yellowstone National Park (YNP). Metabolic reconstruction was coupled with detailed geochemical measurements of each geothermal habitat and reverse-transcriptase PCR to confirm the in situ activity of these populations. Phylogenetic analyses of ribosomal and housekeeping proteins place these archaea near the root of the thaumarchaeal branch. Metabolic reconstruction suggests that these populations are chemoorganotrophic and couple growth with the reduction of oxygen or nitrate in iron oxide habitats, or sulfur in hypoxic sulfur sediments. The iron oxide population has the potential for growth via the oxidation of sulfide to sulfate using a novel reverse sulfate reduction pathway. Possible carbon sources include aromatic compounds (for example, 4-hydroxyphenylacetate), complex carbohydrates (for example, starch), oligopeptides and amino acids. Both populations contain a type III ribulose bisphosphate carboxylase/oxygenase used for carbon dioxide fixation or adenosine monophosphate salvage. No evidence for the oxidation of ammonia was obtained from de novo sequence assemblies. Our results show that thermoacidophilic Thaumarchaeota from oxic iron mats and hypoxic sulfur sediments exhibit different respiratory machinery depending on the presence of oxygen versus sulfide, represent deeply rooted lineages within the phylum Thaumarchaeota and are endemic to numerous sites in YNP.
Similar content being viewed by others
Introduction
The recently proposed archaeal phylum, Thaumarchaeota (Brochier-Armanet et al., 2008a), contains ammonia-oxidizing archaea (AOA) from marine and terrestrial environments. Since the cultivation of the first AOA from this phylum, Nitrosopumilus maritimus (Könneke et al., 2005), two thermophilic AOA have been isolated (Nitrososphaera gargensis, Hatzenpichler et al., 2008; Nitrosocaldus yellowstonii, de la Torre et al., 2008), and these organisms couple nitrification to carbon fixation at temperatures of ∼45 and 75 °C, respectively, and circumneutral to slightly basic pH values (pH ∼7–8). Concurrently, recent studies identified crenarchaeol (a lipid biomarker of AOA) (Pearson et al., 2004) and ammonia monooxygenase subunit A (amoA) genes (Spear et al., 2007; Weidler et al., 2007; Reigstad et al., 2008; Zhang et al., 2008) from geochemically and geographically diverse geothermal systems. These findings raised important questions about the distribution of AOA in thermophilic environments and whether nitrification is an important metabolism in geothermal habitats.
To date, all cultured members of the Thaumarchaeota are autotrophic (via the 3-hydroxypropionate/4-hydroxybutyrate pathway) AOA. Several recent studies have documented the importance of thaumarchaea in nitrification and carbon fixation in marine and terrestrial environments (Pester et al., 2011; Brochier-Armanet et al., 2011a; Hatzenpichler, 2012; Stahl and de la Torre, 2012). However, these organisms are only distantly related to other Thaumarchaeota detected in thermal habitats (Inskeep et al., 2010), and little is known about alternate energy-yielding pathways in this phylum. Planktonic thaumarchaea have been shown to take up amino acids in the ocean (Ouverney and Fuhrman, 2000; Kirchman et al., 2007); however, these measurements cannot directly infer heterotrophic metabolism. The recent isolation of a soil thaumarchaeon, which utilizes pyruvate during nitrification (Tourna et al., 2011), provides evidence that some thaumarchaea can grow mixotrophically. Moreover, recent work shows that amoA-encoding members of the Thaumarchaeota are not obligate autotrophic nitrifiers in wastewater treatment plants and likely utilize organic compounds for growth (Muβmann et al., 2011). The recent genome sequence of a deeply branching thaumarchaeal relative (Brochier-Armanet et al., 2011b) from the proposed phylum ‘Aigarchaeota’ (Candidatus ‘Caldiarchaeum subterraneum’) (Nunoura et al., 2011) does not contain an amoA gene and has provided insights regarding other metabolisms potentially important in the Thaumarchaeota. Furthermore, partial sequence data from a member of the ‘miscellaneous crenarchaeotal group’ suggest that Thaumarchaeota residing in marine sediments degrade detrital proteins in situ (Lloyd et al., 2013). These more recent reports suggest that metabolisms other than ammonia oxidation are likely widespread among members of the Thaumarchaeota.
Prior 16S rRNA gene surveys of iron oxide and elemental sulfur sediment communities in YNP geothermal ecosystems revealed numerous sequences that were distantly related to the Marine Crenarchaeota Group I.1a (Jackson et al., 2001; Inskeep et al., 2004; Macur et al., 2004). The phylogenetic position of these novel archaea had been elusive until metagenome sequencing was used to obtain a greater number of genes corresponding to the thaumarchaeal population from a high-temperature (∼65–68 °C), acidic (pH ∼3) iron oxide mat community, Yellowstone National Park (YNP) (Inskeep et al., 2010). The 16S rRNA gene from this thaumarchaeote is only 84% percent similar (nucleotide identity) to its closest cultivated relative (N. yellowstonii); consequently, it is important to identify the phylogenetic position and metabolic repertoire of novel members of this phylum. Toward this goal, another very recent study described the phylogenetic position of two ‘Hot Thaumarchaeota-related Clades’ (HTC1 and 2) from a hot-spring pool in Kamchatka (Eme et al., 2013). Fosmid sequencing was used to obtain a total of 132 209 bp (HTC1) and 61 236 bp (HTC2) for these thermophilic populations. The small amount of sequence data (<10% of a 1.5-Mb genome) presented in that study makes it difficult to determine the metabolic potential of microorganisms in these clades, as well as their exact relationship with the Thaumarchaeota (based on the presence and absence of thaumarchaeal-specific genes; Spang et al., 2010). Moreover, the geochemical data presented in that study lacked detail sufficient to suggest possible ecological niches of these archaea (for example, dissolved oxygen).
The primary goal of this study was to perform a detailed phylogenetic and functional analysis of two distinct and different thaumarchaeal genome assemblies obtained from random shotgun metagenome sequencing of acidic (pH ∼3), high-temperature (65–72 °C) iron oxide and elemental sulfur sediment communities. Our results show that high-temperature thaumarchaeal populations from both oxic and hypoxic environments likely generate energy via oxidation of organic compounds, and may fix carbon dioxide via a ribulose bisphosphate carboxylase/oxygenase (RuBisCO)-encoding gene thought also to be part of adenosine monophosphate (AMP) salvage in other organisms (Sato et al., 2007). Although electron transport pathways for the two populations were shown to vary in accordance with oxygen content (heme copper oxidase versus bd-ubiquinol-like types), neither possesses the ammonia oxidation genes observed in AOA (for example, amoA). Moreover, detailed phylogenetic analysis using numerous ribosomal and housekeeping proteins available for each population showed that these thermophilic thaumarchaea branch basally relative to the AOA. One of the thaumarchaeal populations presented here forms a novel clade not yet described in the literature.
Materials and methods
Site descriptions
Beowulf and Dragon Springs (Figures 1a and b) (YNP Thermal Inventory IDs NHSP035 and NHSP106, respectively) are located in the One Hundred Springs Plain region of Norris Geyser Basin, Yellowstone National Park (YNP), Wyoming, USA and have been characterized extensively with regard to geochemical, microbiological and physical parameters (Jackson et al., 2001; Langer et al., 2001; Inskeep et al., 2004, 2005; Boyd et al., 2007; D’ Imperio et al., 2007; Inskeep et al., 2010; Kozubal et al., 2012). The source water geochemistry of these two geothermal springs has been quite similar for over a decade of sampling (Ackerman, 2006; Kozubal et al., 2012): both are low-pH (2.9–3.1), high-temperature (72–82 °C), acid–sulfate–chloride springs containing ferrous Fe (40–50 μM), dissolved sulfide (80–120 μM), dissolved hydrogen (10–100 nM) and high levels of dissolved CO2 (2–4 mM dissolved inorganic carbon, which is predominantly H2CO3 at these pH values). Physical and geochemical data for Beowulf Spring (site BE_D) and Dragon Spring (site DE_B) are given in Supplementary Table S1. Both outflow channels exhibit a zone of sulfur deposition followed by Fe(III)-oxide biomineralization (Langner et al., 2001; Inskeep et al., 2004, 2010). The samples discussed in the current study were obtained from the Fe(III) oxide mats within Beowulf Spring and the elemental sulfur deposition zone of Dragon Spring. The sites are ecologically distinct from one another as a result of the inverse relationship between dissolved sulfide (DS) and dissolved oxygen (DO); the sulfur deposition zone of Dragon Spring contained high levels of dissolved sulfide (∼100 μM) and low levels of dissolved oxygen (O2 (aq) <3 μM), whereas the Fe(III) oxide samples from Beowulf Spring were obtained from a zone of oxygenation (O2 (aq) ∼40–60 μM). The metagenome sequence (discussed below) from Beowulf Spring was obtained from samples taken at site BE_D in 2006 (Inskeep et al., 2010), and again in 2010. The Dragon Spring sample was included in the YNP metagenome study as Site 9 (Community Sequencing Program, CSP 79701, Inskeep et al., 2013; Takacs-Vesbach et al., 2013).
Photographs (a, b) of iron oxyhydroxide microbial mats at Beowulf Spring (a) and elemental sulfur sediments at Dragon Spring (b). Arrows indicate approximate sample locations used for geochemical and metagenome analysis (scale bar=20 cm). Nucleotide word frequency-principal component analysis (c, d) of assembled metagenome sequence from iron oxide mats (c, red and teal) and elemental sulfur sediments (d, yellow). The G+C content (%) of contigs (e, f) included in the thaumarchaeal de novo assemblies from Beowulf (red, teal) and Dragon Spring (yellow) is plotted as a function of decreasing scaffold length (x axis). Cumulative scaffold length is shown on the secondary y axis (solid and dotted black lines represent Sanger and 454 data, respectively).
Physical and geochemical measurements
Temperature was measured with a digital thermometer equipped with a T-type thermocouple (Omega Engineering, Inc, Stamford, CT, USA) with an error of ±1 °C. A portable Accumet AP71 pH/mV/°C meter (Fisher Scientific Inc., Waltham, MA, USA) was used to measure pH after calibration in the field using a buffer of pH 4.01 and temperature compensation. Dissolved oxygen was measured using a modified version of the Winkler method. Briefly, a 60-ml syringe filled with geothermal spring water was capped immediately with a septum (zero headspace) to avoid ingassing of atmospheric O2. Concentrated MnSO4 (0.4 ml) and NaN3 (0.4 ml) were added through the septum to preserve dissolved oxygen, which produces a white to yellow flocculent. Addition of concentrated H2SO4 (0.4 ml) dissolved the flocculent, and a sample volume of 30 ml was then titrated with Na2S2O3. Total dissolved sulfide was measured using the amine sulfuric acid method (APHA, 1998a). Aqueous Fe(II) and Fe(III) were measured using the ferrozine method (To et al., 1999). Anions were measured using ion chromatography (Dionex, Sunnyvale, CA). Total soluble metals and other trace elements were determined using inductively coupled plasma optical emission spectroscopy (Perkin Elmer OPTIMA 5300, Waltham, MA, USA). Total soluble ammonium and nitrate were determined using flow injection analysis (Lachat, Hach Co., Loveland, CO, USA) (APHA, 1998b). Dissolved inorganic carbon was measured as previously described (Inskeep et al., 2004, 2010). Solid-phase characterization of Beowulf and Dragon Springs iron oxide and sulfur sediments is described in detail elsewhere (Langner et al., 2001; Inskeep et al., 2004, 2010; Kozubal et al., 2012).
Metagenome sequencing, assembly and annotation
Metagenome sequencing of iron oxide microbial mats from Beowulf Spring was conducted on two different samples taken in 2006 and 2010 using Sanger and 454 sequencing platforms, respectively. Sanger sequence assemblies obtained from 65 °C Fe mats revealed a novel thaumarchaeal population (Inskeep et al., 2010); however, owing to insufficient coverage, additional sequencing was pursued to present a more complete phylogenetic and functional analysis than was previously possible. Consequently, this study includes additional sequencing efforts on samples taken from Beowulf Spring in 2010 and sequenced using 454 sequencing (Department of Energy/Joint Genome Institute, DOE-JGI). This study also focuses on thaumarchaea from sulfur sediment samples taken from Dragon Spring as part of a DOE-JGI Community Sequencing Program in 2007 and subjected to Sanger sequencing. Details of DNA extraction, library construction and subsequent sequencing were discussed previously (Inskeep et al., 2010, 2013; Takacs-Vesbach et al., 2013). Sequence assemblies were obtained using either Celera or Newbler assemblers and submitted for gene calls and annotation using the Integrated Microbial Genome Expert Review (IMG/ER, Markowitz et al., 2012) supported by the DOE-JGI (Walnut Creek, CA, USA). Curated de novo assemblies of thaumarchaeal populations were also gene-called and annotated in integrated microbial genome. To improve assembly of thaumarchaeal sequences, Sanger sequences from the first sample at Beowulf Spring were then used to recruit sequences with high nucleotide identity (>95%) from the 454 metagenome (obtained from the same location), and provided an additional ∼1.2 Mb of thaumarchaeal sequence. Genome completeness was estimated by comparing the amount of nonredundant sequence in each de novo assembly to the genome sizes of sequenced Thaumarchaeota and Ca. ‘C. subterraneum’ (average genome size ∼1.95 Mb), and also by Hidden Markov Model (HMM; Eddy, 2011) searches of archaeal core housekeeping protein-encoding genes (Pfam A database) that are found in at least 90% of all archaeal genomes (Rinke et al., 2013). The curated de novo assemblies are found under the integrated microbial genome submission IDs 1331 (Beowulf, Sanger, Thaumarchaeota archaeon strain BS1), 6991 (Dragon, Sanger, Thaumarchaeota archaeon strain DS1) and 12021 (Beowulf, 454, Thaumarchaeota archaeon strain BS4).
Nucleotide word frequency-principal component analysis
Nucleotide word frequencies (NWFs) (Teeling et al., 2004; Inskeep et al., 2010) created from assembled metagenome sequence were analyzed using principal component analysis (PCA) (online web server, http://gos.jcvi.org/openAccess/scatterPlotViewer2.html) to separate the major taxa contributing to the assemblies from each community. The NWF was set to four, minimum contig size at 1000 bases and the chop sequence size at 1000 bases. Contigs and scaffolds that exhibited similar sequence character (for example, codon usage bias) were separated using NWF-PCA (nucleotide word frequency-principal component analysis) and further screened using G+C content, BLAST assignments and gene annotation. The poor sequence identity of YNP thaumarchaea to other available reference genomes in this group precluded the use of ‘simple’ BLAST analysis as a major tool for identifying thaumarchaeal sequence.
Phylogeny construction
A detailed 16S rRNA gene phylogeny was constructed to determine the exact relationship of the Beowulf and Dragon thaumarchaeal populations to currently cultivated Thaumarchaeota. Sequences retrieved from Genbank were aligned using ClustalW in the MEGA 5.10 software package with default settings (Tamura et al, 2011). The resulting alignment was manually edited and inspected for regions where homology was doubtful and removed. The edited alignment (1448 aligned nucleotides) was utilized to construct a maximum likelihood (ML) phylogenetic tree in MEGA 5.10 and PhyML 3.0 (Guindon et al., 2010; Tamura et al., 2011). The general time-reversible model with four discrete gamma categories and estimated alpha parameter was implemented to construct both ML trees (100 bootstraps).
Phylogenies were also compared using concatenated alignments of numerous single-copy protein-encoding genes. Forty universally conserved ribosomal proteins from the domains Archaea and Eukarya were retrieved from NCBI nr protein database using BLASTp, aligned with MUSCLE in MEGA 5.10 using default settings (Tamura et al., 2011), and concatenated using an open-source Perl script (http://raven.iab.alaska.edu/~ntakebay/teaching/programming/perl-scripts/fastaConcat.pl). The resulting alignment was manually inspected for regions where homology was uncertain and removed. A ML phylogenetic tree was generated using the Jones–Taylor–Thornton model with four discrete gamma categories (Brochier-Armanet et al., 2008a) and estimated alpha parameter (100 bootstraps) in MEGA 5.10 (Tamura et al., 2011). The Le and Gascuel (Le and Gascuel, 2008) model of sequence evolution was also used to generate an ML tree for the concatenated data set of ribosomal proteins in PhyML 3.0 (Guindon et al., 2010) with an estimated alpha parameter and four discrete gamma categories. An ML topoisomerase IB phylogeny was constructed similar to the concatenated ribosomal protein phylogeny from a ClustalW alignment in MEGA 5.10 (Tamura et al., 2011). All other phylogenetic trees were constructed using distance- and parsimony-based methods from ClustalW alignments in MEGA 5.10 (Tamura et al., 2011).
RNA extraction and RT-PCR
Iron oxide (Beowulf Spring) and elemental sulfur sediments (Dragon Spring) were harvested aseptically from the same physicochemical location of the metagenome samples and immediately preserved with RNAlater (Life Technologies Co., Carlsbad, CA, USA). Total RNA was extracted from approximately 500 mg of each sample type using either a freeze/thaw cycle lysis (iron oxide sediments) or mechanical/chemical lysis (sulfur sediments) using the Soil Lysis Solution from the FastRNA Pro Soil-Direct Kit (MP Biomedicals, LLC, Solon, OH, USA). After centrifugation to remove sediment and cellular debris, nucleic acids were extracted in 1 ml of TRI Reagent (Sigma-Aldrich Co., LLC, St Louis, MO, USA) and incubated for 5 min at room temperature. RNase-free chloroform (200 μl) was added to each tube, incubated for 15 min at 25 °C and centrifuged at 12 000 × g for 15 min at 4 °C. After centrifugation, the upper aqueous layer containing nucleic acids was removed and total RNA was then precipitated by the addition of 500 μl of ice-cold, RNase-free 100% 2-propanol and 1 μl of glycogen (Sigma-Aldrich Co., LLC) following incubation at −20 °C overnight. After precipitation, the samples were centrifuged at 13 000 × g for 15 min at 4 °C, followed by decantation and the addition of 750 μl of 70% ethanol, followed by centrifugation as in the previous step. After the pellet dried (∼15 min), it was resuspended in 1 × NEB DNase buffer (New England Biolabs, Inc., Ipswich, MA, USA), and 2 μl (4 units) of NEB DNase I enzyme was added and incubated at 37 °C for 20 min. The DNase I reaction was terminated by the addition of 0.2 volumes of 8 M LiCl and 2.5 volumes of absolute ethanol. After precipitation at −20 °C overnight, the pellet was washed once in 70% ethanol and dried; it was resuspended in Tris-EDTA buffer (10 mM Tris, 1 mM EDTA, pH=7, Life Technologies Co.). Primers designed around the 16S rRNA genes of Group I.1d, I.1e and I.1f thaumarchaea (Figure 2) (ThaumI.1def-F 5′-TAATACCAGCTCCCCGACTG-3′ and ThaumI.1def-R 5′-CTTCGCCACTGTTGGTCTTC-3′) were used to reverse transcribe (RT) 16S rRNA from total RNA extracts using the AccessQuick RT-PCR system (Promega, Corp., Madison, WI, USA) with the following PCR conditions: RT at 45 °C for 45 min, 35 cycles of 94 °C for 2 min, 94 °C for 30 s, annealing at 55 °C for 30 s, extension at 70 °C for 25 s and a final extension at 70 °C for 7 min. RT-PCR products were visualized on a 1.2% ethidium bromide-stained agarose gel. Control reactions containing no RT confirmed the absence of contaminating DNA.
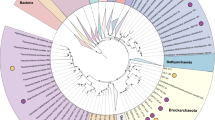
Maximum likelihood 16S rRNA gene tree (1448 unambiguously aligned nucleotides) showing the relationship of the YNP de novo assemblies (bold red) with the archaeal phyla Thaumarchaeota, Crenarchaeota and ‘Aigarchaeota’ rooted with Thermus thermophilus. Filled squares at the nodes indicate bootstrap support by ML in MEGA 5.10 and PhyML >90%. Unfilled squares indicate bootstrap support >70% by ML. Unmarked nodes represent bootstrap support >50%. Scale bar represents the estimated number of substitutions per site. Phylogenetic clades were named following Schleper et al. (2005).
Amplification of amoA genes
Universal primers designed to amplify amoA genes from virtually all environments and groups of AOA (Francis et al., 2005; de la Torre et al., 2008) were used to target amoA genes from community DNA extracts of iron oxide mats (Beowulf Spring) and elemental sulfur sediments (Dragon Spring). Numerous PCR cycling conditions were used from other published studies (Francis et al., 2005; de la Torre et al., 2008; Zhang et al., 2008). Positive controls for archaeal amoA genes were obtained using the primer set from Francis et al. (2005) on soil DNA extracts from the Montana State University A.H. Post Research Farm (Miller et al., 2008).
Ex situ nitrification assay
A 1:1 mixture of iron mat (∼65 °C) from Beowulf Spring and hot spring water was inoculated (∼2 ml) into 150 ml of spring water in sterile 250-ml screw-cap Erlenmeyer flasks (biological triplicates) containing 0.5 mM NH4Cl (∼0.26 nM NH3(aq) at pH=3) and then incubated at 65 °C. Triplicate heat-killed controls were used by boiling the iron mat/spring water mixture on a stove for 15 min. The flasks were manually stirred and opened every 30 min to prevent oxygen limitation. Aqueous samples were obtained at 0, 30, 120 and 220 min using 10 ml of slurry filtered through 0.2-μm nylon filters, transported back to the laboratory (Montana State University) and analyzed for total ammonium and nitrate using flow injection analysis (see above).
Results
De novo thaumarchaeal assemblies
Nucleotide word frequencies (NWFs) (Teeling et al., 2004) of assembled metagenome sequence from an iron oxide mat (Beowulf Spring, Figure 1a) and an elemental sulfur sediment community (Dragon Spring, Figure 1b) were evaluated using principal component analysis to visualize and determine scaffolds of similar sequence character (Figures 1c and d). Each site contained significant assemblies corresponding to unique thaumarchaeal populations that were separated from other community members using NWF-PCA (Figures 1c and d). After further screening using G+C content (%) and scaffold coverage, confident assignment of scaffolds to these thaumarchaeal populations resulted in de novo assemblies of ∼1.2–1.5 Mb from each site (Figures 1e and f; Table 1), which represent 15% (Sanger), 4% (454) and 22.5% (Sanger) of the total metagenome sequence reads from Beowulf and Dragon Springs, respectively. Assemblies from Beowulf Spring (Sanger and 454) exhibited slightly higher G+C content (45.6±2.0 and 42.2±2.4) compared with those from Dragon Spring (40.3±2.0) (Figures 1e and f, Table 1). An inventory of conserved archaeal core protein-encoding genes (Rinke et al., 2013) suggested a genome completeness of ∼80 and 90% for the Beowulf and Dragon thaumarchaeal populations, respectively. Moreover, nearly all tRNA synthetases are present for each thaumarchaeal population type (17/20 and 19/20 for the Beowulf and Dragon assemblies, respectively). The lower number of tRNAs identified in the Beowulf assemblies (Table 1) is likely due to the presence of non-canonical tRNA introns, similar to N. gargensis (Spang et al., 2012). The large amount of nonredundant genome sequence for each of these population types also provided an opportunity to search for thaumarchaeal-specific genes (Spang et al., 2010). Most of the genes present in sequenced Thaumarchaeota (that is, AOA) are conserved in these thermoacidophilic thaumarchaea (Supplementary Table S2) and likely represent synapomorphic traits shared by all members of this phylum.
Phylogenetic analysis
A detailed and thorough 16S rRNA gene phylogeny was constructed using full-length sequences from the YNP metagenome assemblies, as well as sequences from numerous YNP geothermal springs contributed by our research group over the past decade, thaumarchaeal isolates, entries from other geothermal environments and the recently described ‘Hot Thaumarchaeota-related Clade’ 1 and 2 (Eme et al., 2013). Three new phylogenetic clades (Groups I.1d, I.1e and I.1f) are evident from our phylogenetic analysis (Figure 2). Group I.1d exclusively comprises sequences we have obtained from iron oxide mats of acidic hot springs in YNP (Beowulf and Rainbow (RS3) Springs), whereas Group I.1e consists of sequences from both iron mats (that is, Whirligig and Echinus Geysers) and elemental sulfur sediments (that is, Dragon Spring, Joseph’s Coat Hot Springs, and Monarch Geyser). The third and most deeply rooted group of the Thaumarchaeota (I.1f) contains sequences from higher-pH (5.5–6.5) hypoxic springs (that is, Joseph’s Coat and Sylvan Springs). The recent entries from Kamchatka (Eme et al., 2013) fall in Groups I.1e and I.1f. These three new thaumarchaeal clades are significantly different from one another (<84% nucleotide identities), as well as from current thaumarchaeal relatives (for example, ∼85–86% nucleotide identity to N. yellowstonii), and likely represent order-level taxonomic lineages within this phylum. To verify the presence and activity of these populations in the same habitats (Beowulf and Dragon Springs) from which metagenome assemblies were obtained, we performed RT-PCR of 16S rRNA using primers designed to capture Group I.1d, I.1e and I.1f populations from environmental RNA (Supplementary Figure S1a and b). Ribosomal RNA transcription from Group I.1d and I.1e Thaumarchaeota was confirmed in Beowulf and Dragon Springs, respectively.
A concatenated alignment of 40 universally conserved ribosomal proteins (5563 unambiguously aligned amino-acid positions) from the YNP thaumarchaeal de novo genome assemblies was used to construct an ML phylogenetic tree. The organisms from Beowulf and Dragon Springs branch deeper than any currently known Thaumarchaeota (Figure 3). The thaumarchaea referred to as ‘Hot Thaumarchaeota-related Clade’ 1 and 2 cannot be included in this tree, owing to insufficient sequence; however, we would expect these entries to remain within Group I.1e, as suggested by 16S rRNA gene phylogeny. All Thaumarchaeota possess a topoisomerase IB, which has not been identified in other archaea (Brochier-Armanet et al., 2008b). An inconsistent topology with the concatenated ribosomal protein tree was observed with topoisomerase IB sequences from Beowulf and Dragon assemblies; however, their basal position was retained compared with the AOA (Figure 4). The thaumarchaeal topoisomerase IBs form a sister group with the Eukarya, consistent with prior work (Brochier-Armanet et al., 2008b). Other phylogenetic marker protein sequences (that is, PCNA homologs, DNA polymerase small subunit D and RNA polymerase large subunit A) were used to confirm the phylogenetic position of high-temperature thaumarchaea from YNP (Supplementary Figures S2a–c). All phylogenetic trees placed the thermophilic thaumarchaea from acidic iron and sulfur environments in YNP among the most deeply rooted members of the domain Archaea. These observations support the hypothesis that these organisms represent extant relatives of ancestral Thaumarchaeota, which existed before the radiation of low[er]-temperature marine and terrestrial thaumarchaea (Figure 3).
Maximum likelihood phylogenetic tree of 40 concatenated universally conserved ribosomal proteins (5563 unambiguously aligned amino-acid positions) from the de novo YNP thaumarchaeal assemblies (bold red) compared with representatives of the Archaea and Eukarya. Numbers at nodes represent bootstrap values from 100 replications implemented in MEGA 5.10 and PhyML 3.0 (dashes indicate <50% bootstrap support in PhyML), respectively; numbers in parentheses represent the number of taxa included in each collapsed node. Scale bar indicates estimated number of substitutions per site.
Unrooted topoisomerase IB phylogeny of the phylum Thaumarchaeota, Ca. ‘C. subterraneum’, Nitrosocaldus-like and the domain Eukarya. Full-length (∼530 deduced amino acids) topoisomerase IB sequences from the de novo assemblies (red bold) are shown relative to other thaumarchaeal isolates (bold black). Numbers at the nodes represent bootstrap values from 100 replications implemented in MEGA 5.10 and PhyML 3.0, respectively. Scale bar indicates estimated number of substitutions per site.
Energy metabolism
A detailed metabolic model was created for both de novo assemblies, which highlights the differences and similarities between the Beowulf and Dragon thaumarchaeal populations (Figure 5; Supplementary Table S2). The difference in dissolved oxygen concentration between the iron oxide mats in Beowulf Spring (O2 (aq)∼40 μM) and the sulfur sediments of Dragon Spring (O2 (aq)<3 μM) (Supplementary Table S1) was reflected in important attributes of each population. Heme copper (terminal) oxidases (HCOs) were present in iron oxide thaumarchaea but absent in populations from hypoxic sulfur sediments. In addition to HCOs, several blue copper proteins were identified in the iron oxide assemblies, some of which are present adjacent to HCO operons. Blue copper proteins were not present in populations from hypoxic sulfur communities. Conversely, the thaumarchaea from Dragon Spring contained the A subunit of the high-affinity cytochrome bd-type terminal oxidase (cydA), as well as a duplicated A subunit (cydA′). However, the gene encoding the oxygen-binding subunit (cydB) (Junemann, 1997) is absent. Without an oxygen-binding subunit, this thaumarchaeal population likely does not respire oxygen. Both populations contained genes encoding for superoxide dismutase (sodA) and alkyl hydroperoxide reductase (ahp), which are required for detoxification of reactive oxygen species (ROSs). The thaumarchaeal population from Dragon Spring also contained genes necessary to reduce elemental sulfur to hydrogen sulfide, similar to the pathway in Pyrococcus furiosus (Ma and Adams, 1994; Hagen et al., 2000; Bridger et al., 2011). Genes that encode sulfide dehydrogenase (sudBA), a partial MBX-like NADPH dehydrogenase (mbx), and NADPH:sulfur oxidoreductase (nsr) were all present in the Dragon assemblies. Moreover, the consensus sequence from Dragon Spring revealed metabolic potential to ferment acetate via a gene encoding for an ADP-forming acetyl-CoA synthetase. Conversely, the Beowulf Spring assemblies contained a complete membrane-bound nitrate reductase (narGHJI) operon (Martinez-Espinosa et al., 2007), which indicates that this population has the potential to grow anaerobically by reducing nitrate to nitrite. However, the absence of nitrite reductase (nir) and nitric oxide reductase (nor) genes in both populations suggests that they are not capable of complete denitrification.
Metabolic reconstruction of energy generation, central carbon metabolism and general transport functions deduced from the Beowulf (red) and Dragon (yellow) genome sequence assemblies. Shared functions and intermediates are shown in either black text or filled blue boxes. Reducing equivalents (for example, NADH and ferredoxin) produced in central carbon pathways are fed into the respiratory chain at the NADH-ubiquinone oxidoreductase complex (NUO). Alternatively, the Dragon population may utilize reduced ferredoxin to reduce NADP+ via a MBX-like NADPH dehydrogenase (MBX), which then can reduce elemental sulfur by either NADPH: sulfur oxidoreductase (NSR) or sulfide dehydrogenase (SuDH). The sequence from the Beowulf population indicates that reducing equivalents can be produced during sulfide oxidation by sulfide dehydrogenase (SuDH), a hypothetical heterodisulfide-like enzyme complex, adenosine-5′-phosphosulfate reductase (APS) and sulfate adenylyltransferase (SAT) coupled to either the reduction of oxygen by heme copper oxidases (HCO) or nitrate by a nitrate reductase (NAR) complex. Both populations contain genes for energy generation via succinate dehydrogenase (II). Red and yellow dashed lines indicate possible electron flow for the Beowulf and Dragon populations, respectively.
The thermophilic thaumarchaeal populations from iron oxide and sulfur sediment communities contained numerous genes coding for proteins involved in chemoorganotrophic growth on complex carbohydrates, peptides, amino acids and simple organic compounds. For example, each assembly contained genes for alpha-amylases and glycoside hydrolases, which encode for enzymes that degrade starch into glucose and maltose to be used in glycolysis (via glucokinase). Multiple peptidases and amino-acid and peptide transporters were also found in each population. In addition, the Group I.1d population (iron oxide mats) contained protein-encoding genes for the catabolism of aromatic compounds (for example, 4-hydroxyphenylacetate, 4-HPA). The 4-hydroxyphenylacetate-3-monooxygenase-encoding gene converts 4-hydroxyphenylacetate to protocatchuate, which can be incorporated into the tricarboxylic acid (TCA) cycle via succinate and pyruvate conversion to acetyl-CoA by pyruvate:ferredoxin oxidoreductase (Fuchs et al., 2011).
The Group I.1d population from iron oxide mats also contained protein-encoding genes known to be responsible for lithotrophic growth on reduced sulfur compounds (for example, H2S) via a novel reverse sulfate reduction pathway. Specifically, dissimilatory adenosine-5′-phosphosulfate reductase (aprBA), sulfate adenylyltransferase (sat), flavoprotein sulfide dehydrogenase (sudH) and a drsE-like gene are found together in an operon-like region that also contains an SirA-like transcriptional regulator (Supplementary Figure S3a; Quatrini et al., 2009). The AprA sequence from the Beowulf thaumarchaeal population is phylogenetically distinct from any sequence currently in the NCBI nr database, and branches basally to all sulfur-oxidizing bacterial AprAs used in the reverse sulfate reduction pathway (Supplementary Figure S3b). Importantly, both thaumarchaeal populations from YNP lack genes coding for amoA, X, C or B, which are observed in AOA (Supplementary Table S2; Walker et al., 2010). Moreover, no amoA genes were PCR amplified from environmental DNA extracts from either geothermal site using primers designed to capture amoA genes from diverse environments (Francis et al., 2005; de la Torre et al., 2008). Furthermore, a prior metagenome study also failed to detect any amoA genes in high-temperature iron oxide mats (Inskeep et al., 2010, 2013). Finally, no oxidation of ammonia was observed in ex situ bioassays using live iron oxide microbial mats from Beowulf Spring. No change in either total ammonium or nitrate was observed in triplicate vessels (Supplementary Figure S4). Similar experiments were not conducted using sulfur sediments from Dragon Spring, because high levels of hydrogen sulfide (>100 μM) are known to inhibit ammonia oxidation (for example, Joye and Hollibaugh, 1995). Although significant amounts of ammonium (∼60 μM; Supplementary Table S1) are present in these geothermal springs, the high sulfide levels (Dragon) and low pH (∼3) at both sites likely exclude ammonia-oxidizing metabolisms. The substrate for ammonia monooxygenase is ammonia (Suzuki et al., 1974) and would be predicted to be ∼0.04 nM at a pH of ∼3. The lowest reported ammonia concentration supporting growth is 0.19 nM for the acidophilic AOA Ca. ‘N. devanaterra’ (Lehtovirta-Morley et al., 2011). Collectively, the genomic and geochemical data strongly suggest that ammonia oxidation is absent in these thermoacidophilic Thaumarchaeota.
Central carbon metabolism
Several cultivated Thaumarchaeota contain the 3-hydroxypropionate/4-hydroxybutyrate carbon fixation pathway first identified in Metallosphaera sedula (Berg et al., 2007). However, the potential for carbon dioxide fixation by the YNP thaumarchaeal populations is not conclusive from the current de novo assemblies. For example, both YNP populations lack genes coding for the key enzyme of this pathway (4-hydroxybutyryl-CoA dehydratase, hcd), which is consistent with the absence of this gene in Ca. ‘C. subterraneum’ (Nunoura et al., 2011). However, the Beowulf population contained genes coding for the vitamin B12-dependent enzyme, methylmalonyl-CoA mutase (mcm), and the bifunctional, biotin-dependent, acetyl/propionyl-CoA carboxylase (accABC). Although accABC genes were absent from the Dragon assembly, this population contained copies of phosphoenolpyruvate carboxylase and a pyruvate:water dikinase; both genes encode enzymes involved in the dicarboxylate/4-hydroxybutyrate pathway described in Ignicococcus hospitalis (Huber et al., 2008). Consequently, although each population contained genes suggestive of possible autotrophic pathways, a definitive case for autotrophy based on known pathways cannot be made from the current sequence data. Characterization of cultivated strains relevant to these systems will likely be necessary to resolve the potential for autotrophic fixation of carbon dioxide in these populations, and to determine whether this may occur via novel pathways.
Both YNP populations contained all genes required for the Embden–Meyerhof and semi-phosphorylative Entner–Doudoroff pathways (Siebers and Schonheit, 2005). Fructose-1, 6-bisphosphotase was also identified, and it provides a mechanism for incorporating carbon into cellular biomass when growing on amino acids, peptides or tricarboxylic acid intermediates. All protein-encoding genes for a complete tricarboxylic acid cycle were present in the Beowulf thaumarchaea. The Dragon population possibly contains a branched tricarboxylic acid cycle, which is a common attribute of fermenting or anaerobic microorganisms. Pentose phosphate sugars for nucleotide synthesis are likely formed by a reverse ribulose monophosphate (RuMP) pathway, as indicated by genes encoding for a 3-hexulose-6-phosphate synthase and 6-phospho-3-hexuloisomerase in the Dragon assembly (Soderberg, 2005). The Beowulf population lacks all genes for a reverse ribulose monophosphate pathway, but contains genes encoding for a putative F420-dependent glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase. The presence of these genes indicate potential for an oxidative pentose phosphate pathway; similar to N. gargensis (Spang et al., 2012), the Beowulf population lacks a 6-phosphoglucono-δ-lactonase gene.
A Type III RuBisCO is present in both thermoacidophilic thaumarchaea from YNP, similar to those found in some Desulfurococcales and Euryarchaeota (Sato et al., 2007). Although not identified previously in any known Thaumarchaeota, this type of RuBisCO may be used in an AMP salvage pathway along with an AMP-phosphorylase (DeoA) and ribose-1, 5-bisphosphate isomerase (E2B1) to yield 3-phosphoglycerate (Sato et al., 2007). The YNP thaumarchaeal assemblies do not contain any ADP-dependent sugar kinases known to result in significant accumulation of intracellular AMP. However, they both contain AMP-forming acyl-CoA synthetases that could produce high intracellular AMP concentrations. Consequently, RuBisCO may participate in an alternative carbon fixation pathway or AMP salvage in these thermophilic Thaumarchaeota. Moreover, the presence of a RuBisCO gene in these thaumarchaeal populations provides a potential evolutionary link between RuBisCO genes observed in other Archaea (Tabita et al., 2008).
Cell division and replication
Most characterized crenarchaea and thaumarchaea use a set of cell division proteins (CdvABC) that have also been characterized in numerous Eukarya as part of the Endosomal Sorting Complex Required for Transport (ESCRT)-III system (Samson et al., 2008; Makarova et al., 2010; Pelve et al., 2011). Both thaumarchaeal assemblies from YNP have a cdvBBC operon and another cdvB gene located elsewhere; however, no cdvA gene was identified (Supplementary Table S2). The cdvA gene is also absent from Ca. ‘C. subterraneum’, yet this organism contains ftsZ (Nunoura et al., 2011), and members of the Euryarchaeota are known to use FtsZ proteins in cell division (Makarova et al., 2010). Although no ftsZ genes were present in either of the YNP thaumarchaeal populations, sepF orthologs were present, and SepF has been proposed to interact with FtsZ during cellular division in members of the Euryarchaeota (Supplementary Table S2; Makarova et al., 2010). Moreover, neither de novo assembly contains actin or artubulin homologs (Supplementary Table S2) present in other archaea (Yutin and Koonin, 2012). Consequently, details regarding cellular division mechanisms in these thermophilic thaumarchaeal populations are not entirely resolvable from genome assemblies. Finally, both of the YNP populations encode for two orc1/cdc6 genes similar to N. gargensis (Spang et al., 2012), which suggests two separate origins of replication (oriC), as cdc6 genes are often located near the origins of replication (Gaudier et al., 2007).
Discussion
This is the first detailed genomic, phylogenetic and metabolic analysis of two different novel thermoacidophilic thaumarchaeal populations found in iron oxide and sulfur sediment microbial communities in YNP. Although these thaumarchaea have thus far eluded cultivation, the de novo assemblies provide insight regarding possible carbon sources and mechanisms of energy conservation. The thermophilic Thaumarchaeota from YNP have metabolic capabilities consistent with their respective geochemical environments, which provide several options for growth in these microbial communities. Analysis of the Beowulf thaumarchaeal population revealed a significant metabolic repertoire contained within a relatively small genome (estimated size ∼1.7 Mb). Multiple HCOs likely allow this population to function under varying O2 and electron donor availability. This organism can also potentially use nitrate as a terminal electron acceptor under anaerobic conditions coupled to the oxidation of sulfide and/or organic compounds. This may be an important metabolic capability under hypoxic conditions known to occur below depths of ∼0.7 mm in iron oxide microbial mats of the same systems (Bernstein et al., 2013). The fate of nitrite potentially produced by these populations is unclear, as no obvious modes of nitrite oxidation or reduction are apparent in any member of these communities (for example, nir gene family) (Inskeep et al., 2010, 2013). The thaumarchaeal population from Beowulf Spring contains the most deeply rooted AprA protein (Supplementary Figure S3b) currently known, which is used to oxidize reduced sulfur compounds using a novel reverse sulfate reduction pathway (Figure 5), and is common to many bathypelagic (for example, SUP05) and mussel symbiont (for example, Bathymodiolus spp.) sulfur-oxidizing bacteria (Walsh et al., 2009). This is the first evidence of a novel reverse sulfate reduction pathway in the domain Archaea, which may represent an evolutionary link between bathypelagic and symbiont bacterial sulfur oxidation pathways and the sulfate reduction pathways of the thermophilic Thermoproteales (Supplementary Figure S3b).
The thaumarchaeal population from Dragon Spring represents the most deeply rooted member of the Thaumarchaeota, and this is the first organism within this phylum to lack HCOs. The lack of HCOs and the presence of bd-ubiquinol-like oxidases in the Dragon population is consistent with the absence of oxygen (and high dissolved sulfide) in this environment and may provide clues regarding the evolution of aerobic metabolism in the Thaumarchaeota. Protein-coding genes in the Dragon population responsible for starch degradation coupled with the reduction of elemental sulfur, and for acetate fermentation, are consistent with important metabolic processes typical of an anoxic habitat replete with sulfide and elemental sulfur. The concentrations of dissolved inorganic carbon within the outflow channels of Dragon and Beowulf Springs range from ∼0.5 to 2 mM (Supplementary Table S1). The large majority of dissolved inorganic carbon is present as CO2 (aq) (that is, H2CO3) at the spring water pH values near 3. Consequently, both of these thaumarchaeal populations are found in environments where aqueous CO2 levels are ∼100 times oversaturated with respect to current atmospheric levels of CO2 (g) (that is, 0.0004 atm) (Inskeep et al., 2005). Genome assemblies of both populations provide evidence that these archaea have the metabolic potential to fix dissolved inorganic carbon to 3-phosphoglycerate (Figure 5) via a novel RuBisCO pathway. To date, this alternative to the classical Calvin–Benson–Bassham cycle has only been identified in select members of the Euryarchaeota (for example, Thermococcales) (Sato et al., 2007). The presence of this set of genes in representatives of the Thaumarchaeota is another exciting area of further research to confirm the function and possible origin of proteins not only found in other organisms but also critical to their adaptive radiation (that is, RuBisCO).
It is becoming evident that members of the Thaumarchaeota are among the most diverse of all known phyla within the domain Archaea, and that their evolutionary adaptations have resulted in an extensive radiation into numerous different habitat types. Phylogenetic and functional genomic analyses of the thaumarchaeal lineages discussed in this study provide evidence that alternate mechanisms of energy generation are available to these organisms besides the oxidation of ammonia. The thermoacidophilic Thaumarchaeota inhabiting iron oxide and elemental sulfur sediment communities do not contain ammonia monooxygenase genes (for example, amoA) observed in AOA (Supplementary Table S2). Moreover, amoA genes have not been identified in numerous geothermal habitats similar to those discussed here (Inskeep et al., 2010, 2013), nor could they be amplified from DNA extracts from these microbial communities. Evidence for complete chemoorganotrophic pathways, chemolithotrophic energy sources and differential respiratory pathways in the two populations are consistent with the geochemical attributes of these disparate habitat types. The fact that these two thermoacidophilic lineages branch basally relative to other known ammonia-oxidizing thaumarchaea supports the hypothesis that ammonia oxidation is a derived physiologic trait in the phylum Thaumarchaeota. Genomes from other thaumarchaea that reside in marine sediments (for example, pSL12 group), soils and natural waters, as well as other thermophilic habitats (for example, Group I.1f and Nitrosocaldus-like), will undoubtedly provide additional insight regarding the diversity of physiological attributes within members of this important and ubiquitous phylum.
References
Ackerman GG . (2006), Biogeochemical gradients and energetics in geothermal systems of Yellowstone National Park. MSc thesis, Montana State University: Bozeman, MT, USA, p 196.
APHA (1998a). Part 4500: S2− D. In: Clesceri LS, Greenberg AE, Eaton AD (eds) Standard Methods for the Examination of Water and Wastewater 20th edn. APHA: Washington, DC, USA, pp 4-165–166.
APHA (1998b). Part 4500: NH3 H. In: Clesceri LS, Greenberg AE, Eaton AD (eds) Standard Methods for the Examination of Water and Wastewater 20th edn. APHA: Washington, DC, USA, pp 4-111–112.
APHA (1998c). Part 4500: NO3− I. In: Clesceri LS, Greenberg AE, Eaton AD (eds) Standard Methods for the Examination of Water and Wastewater 20th edn. APHA: Washington, DC, USA, pp. 4-121–122.
Berg I, Kockelkorn D, Buckel W, Fuchs G . (2007). A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in archaea. Science 318: 1782–1786.
Bernstein HC, Beam JP, Kozubal MA, Carlson RP, Inskeep WP . (2013). In situ analysis of oxygen consumption and diffusive transport in high-temperature acidic iron-oxide microbial mats. Environ Microbiol 15: 2360–2370.
Boyd E, Jackson R, Encarnacion G, Zahn J, Beard T, Leavitt W et al (2007). Isolation, characterization, and ecology of sulfur-respiring Crenarchaea inhabiting acid-sulfate-chloride-containing geothermal springs in Yellowstone National Park. ApplEnviron Microbiol 73: 6669–6677.
Bridger S, Clarkson S, Stirrett K, DeBarry M, Lipscomb G, Schut G et al (2011). Deletion strains reveal metabolic roles for key elemental sulfur-responsive proteins in Pyrococcus furiosus. JBacteriol 193: 6498–6504.
Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P . (2008a). Mesophilic crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol 6: 245–252.
Brochier-Armanet C, Forterre P, Gribaldo S . (2011b). Phylogeny and evolution of the Archaea: one hundred genomes later. Curr Opin Microbiol 14: 274–281.
Brochier-Armanet C, Gribaldo S, Forterre P . (2008b). A DNA topoisomerase IB in Thaumarchaeota testifies for the presence of this enzyme in the last common ancestor of Archaea and Eucarya. Biol Direct 3: 54.
Brochier-Armanet C, Gribaldo S, Forterre P . (2011a). Spotlight on the Thaumarchaeota. ISME J 6: 227–230.
de la Torre JR, Walker CB, Ingalls AE, Könneke M, Stahl DA . (2008). Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ Microbiol 10: 810–818.
D’ Imperio S, Lehr C, Breary M, McDermott T . (2007). Autecology of an arsenite chemolithotroph: sulfide constraints on function and distribution in a geothermal spring. Appl Environ Microbiol 73: 7067–7074.
Eddy S . (2011). Accelerated profile HMM searches. PLoS Comput Biol 7: e1002195.
Eme L, Reigstad LJ, Spang A, Lanzén A, Weinmaier T, Rattei T et al (2013). Metagenomics of Kamchatkan hot spring filaments reveal two new major (hyper)thermophilic lineages related to Thaumarchaeota. Res Microbiol 164: 425–438.
Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB . (2005). Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA 102: 14683–14688.
Fuchs G, Boll M, Heider J . (2011). Microbial degradation of aromatic compounds—from one strategy to four. Nat Rev Microbiol 9: 803–816.
Gaudier M, Schuwirth BS, Westcott SL, Wigley DB . (2007). Structural basis of DNA replication origin recognition by an ORC protein. Science 317: 1213–1216.
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O . (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321.
Hagen W, Silva P, Amorim M, Hagedoorn P, Wassink H, Haaker H et al (2000). Novel structure and redox chemistry of the prosthetic groups of the iron-sulfur flavoprotein sulfide dehydrogenase from Pyrococcus furiosus; evidence for a [2Fe-2S] cluster with Asp(Cys)(3) ligands. J Biol Inorg Chem 5: 527–534.
Hatzenpichler R . (2012). Diversity, physiology, and niche differentiation of ammonia-oxidizing archaea. Appl Environ Microbiol 78: 7501–7510.
Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H et al (2008). A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci USA 105: 2134–2139.
Huber H, Gallenberger M, Jahn U, Eylert E, Berg I, Kockelkorn D et al (2008). A dicarboxylate/4-hydroxybutyrate autotrophic carbon assimilation cycle in the hyperthermophilic archaeum Ignicoccus hospitalis. Proc Natl Acad Sci USA 105: 7851–7856.
Inskeep W, Ackerman G, Taylor W, Kozubal M, Korf S, Macur R . (2005). On the energetics of chemolithotrophy in nonequilibrium systems: case studies of geothermal springs in Yellowstone National Park. Geobiology 3: 297–317.
Inskeep W, Macur R, Harrison G, Bostick B, Fendorf S . (2004). Biomineralization of As(V)-hydrous ferric oxyhydroxide in microbial mats of an acid-sulfate-chloride geothermal spring, Yellowstone National Park. Geochim Cosmochim Acta 68: 3141–3155.
Inskeep W, Rusch D, Jay Z, Herrgard M, Kozubal M, Richardson T et al (2010). Metagenomes from high-temperature chemotrophic systems reveal geochemical controls on microbial community structure and function. PLoS One 5: e9773.
Inskeep WP, Jay ZJ, Herrgard MJ, Kozubal MA, Rusch DB, Tringe SG et al (2013). Phylogenetic and functional analysis of metagenome sequence from high-temperature archaeal habitats demonstrate linkages between metabolic potential and geochemistry. Front Microbiol 4: 95.
Inskeep WP, Jay ZJ, Tringe SG, Herrgard M, Rusch DB et al (2013). The YNP metagenome project: environmental parameters responsible for microbial distribution in the Yellowstone geothermal ecosystem. Front Microbiol 4: 67.
Jackson C, Langner H, Donahoe-Christiansen J, Inskeep W, McDermott T . (2001). Molecular analysis of microbial community structure in an arsenite-oxidizing acidic thermal spring. Environ Microbiol 3: 532–542.
Joye S, Hollibaugh J . (1995). Influence of sulfide inhibition of nitrification on nitrogen regeneration in sediments. Science 270: 623–625.
Junemann S . (1997). Cytochrome bd terminal oxidase. Biochim Biophys Acta Bioenerg 1321: 107–127.
Kirchman DL, Elifantz H, Dittel AI, Malmstrom RR, Cottrell MT . (2007). Standing stocks and activity of archaea and bacteria in the western arctic ocean. Limnol Oceanogr 52: 495–507.
Kozubal MA, Macur RE, Jay ZJ, Beam JP, Malfatti SA, Tringe SG et al (2012). Microbial iron cycling in acidic geothermal springs of Yellowstone National Park: integrating molecular surveys, geochemical processes, and isolation of novel Fe-active microorganisms. Front Microbiol 3: 109.
Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA . (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437: 543–546.
Langner H, Jackson C, McDermott T, Inskeep W . (2001). Rapid oxidation of arsenite in a hot spring ecosystem, Yellowstone National Park. Environ SciTechnol 35: 3302–3309.
Le SQ, Gascuel O . (2008). An improved general amino acid replacement matrix. Mol Biol Evol 25: 1307–1320.
Lehtovirta-Morley L, Stoecker K, Vilcinskas A, Prosser J, Nicol G . (2011). Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc Natl Acad Sci USA 108: 15892–15897.
Lloyd KG, Schreiber L, Peterson DG, Kjeldsen KU, Lever MA, Steen AD et al (2013). Predominant archaea in marine sediments degrade detrital proteins. Nature 496: 215–218.
Ma K, Adams M . (1994). Sulfide dehydrogenase from the hyperthermophilic archaon Pyrococcus furiosus—a new multifunctional enzyme involved in the reduction of elemental sulfur. J Bacteriol 176: 6509–6517.
Macur R, Langner H, Kocar B, Inskeep W . (2004). Linking geochemical processes with microbial community analysis: successional dynamics in an arsenic-rich, acid-sulphate-chloride geothermal spring. Geobiology 2: 163–177.
Makarova K, Yutin N, Bell S, Koonin E . (2010). Evolution of diverse cell division and vesicle formation systems in archaea. Nat Rev Microbiol 8: 731–741.
Markowitz V, Chen I-M, Palaniappan K, Chu K, Szeto E, Grechkin Y et al (2012). IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res 40: D115–D122.
Martinez-Espinosa R, Dridge E, Bonete M, Butt J, Butler C, Sargent F et al (2007). Look on the positive side! The orientation, identification and bioenergetics of ‘archaeal’ membrane-bound nitrate reductases. FEMS Microbiol Lett 276: 129–139.
Miller P, Buschena D, Jones C, Holmes J . (2008). Transition from intensive tillage to no-tillage and organic diversified annual cropping systems. Agron J 100: 591–599.
Muβmann M, Brito I, Pitcher A, Damste J, Hatzenpichler R, Richter A et al (2011). Thaumarchaeotes abundant in refinery nitrifying sludges express amoA but are not obligate autotrophic ammonia oxidizers. Proc Natl Acad Sci USA 108: 16771–16776.
Nunoura T, Takaki Y, Kakuta J, Nishi S, Sugahara J, Kazama H et al (2011). Insights into the evolution of archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res 39: 3204–3223.
Ouverney CC, Fuhrman JA . (2000). Marine planktonic archaea take up amino acids. Appl Environ Microbiol 66: 4829–4833.
Pearson A, Huang Z, Ingalls A, Romanek C, Wiegel J, Freeman K et al (2004). Nonmarine crenarchaeol in Nevada hot springs. Appl Environ Microbiol 70: 5229–5237.
Pelve E, Lindas A, Martens-Habbena W, de la Torre J, Stahl D, Bernander R . (2011). Cdv-based cell division and cell cycle organization in the thaumarchaeon Nitrosopumilus maritimus. Mol Microbiol 82: 555–566.
Pester M, Schleper C, Wagner M . (2011). The Thaumarchaeota: an emerging view of their phylogeny and ecophysiology. Curr Opin Microbiol 14: 1–7.
Quatrini R, Appia-Ayme C, Denis Y, Jedlicki E, Holmes DS, Bonnefoy V . (2009). Extending the models for iron and sulfur oxidation in the extreme acidophile Acidithiobacillus ferrooxidans. BMC Genomics 10: 394.
Reigstad LJ, Richter A, Daims H, Urich T, Schwark L, Schleper C . (2008). Nitrification in terrestrial hot springs of Iceland and Kamchatka. FEMS Microbiol Ecol 64: 167–174.
Rinke C, Schwientek P, Sczyrba A, Ivanova N, Anderson I, Cheng J-F et al (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature 499: 431–437.
Samson R, Obita T, Freund S, Williams R, Bell S . (2008). A role for the ESCRT system in cell division in archaea. Science 322: 1710–1713.
Sato T, Atomi H, Imanaka T . (2007). Archaeal type III RuBisCOs function in a pathway for AMP metabolism. Science 315: 1003–1006.
Schleper C, Jurgens G, Jonuscheit M . (2005). Genomic studies of uncultivated archaea. Nat Rev Microbiol 3: 479–488.
Siebers B, Schonheit P . (2005). Unusual pathways and enzymes of central carbohydrate metabolism in archaea. Curr Opin Microbiol 8: 695–705.
Soderberg T . (2005). Biosynthesis of ribose-5-phophate and erythrose-4-phosphate in archaea: a phylogenetic analysis of archaeal genomes. Archaea 1: 347–352.
Spang A, Hatzenpichler R, Brochier-Armanet C, Rattei T, Tischler P, Spieck E et al (2010). Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol 18: 331–340.
Spang A, Poehlein A, Offre P, Zumbrägel S, Haider S, Rychlik N et al (2012). The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: insights into metabolic versatility and environmental adaptations. Environ Microbiol 14: 3122–3145.
Spear J, Barton H, Robertson C, Francis C, Pace N . (2007). Microbial community biofabrics in a geothermal mine adit. Appl Environ Microbiol 73: 6172–6180.
Stahl DA, de la Torre JR . (2012). Physiology and diversity of ammonia-oxidizing archaea. Annu Rev Microbiol 66: 83–101.
Suzuki I, Dular U, Kwok SC . (1974). Ammonia or ammonium ion as substrate for oxidation by Nitrosomonas europaea cells and extracts. J Bacteriol 120: 556–558.
Tabita F, Hanson T, Satagopan S, Witte B, Kreel N . (2008). Phylogenetic and evolutionary relationships of RubisCO and the RubisCO-like proteins and the functional lessons provided by diverse molecular forms. Philos Trans R Soc B Biol Sci 363: 2629–2640.
Takacs-Vesbach C, Inskeep WP, Jay ZJ, Herrgard M, Rusch DB, Tringe SG et al (2013). Metagenome sequence analysis of filamentous microbial communities obtained from geochemically distinct geothermal channels reveals specialization of three aquificales lineages. Front Microbiol 4: 84.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S . (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739.
Teeling H, Meyerdierks A, Bauer M, Amann R, Glockner F . (2004). Application of tetranucleotide frequencies for the assignment of genomic fragments. Environ Microbiol 6: 938–947.
To T, Nordstrom D, Cunningham K, Ball J, McCleskey R . (1999). New method for the direct determination of dissolved Fe(III) concentration in acid mine waters. Environ Sci Technol 33: 807–813.
Tourna M, Stieglmeier M, Spang A, Könneke M, Schintlmeister A, Urich T et al (2011). Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci USA 108: 8420–8425.
Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ et al (2010). Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA 107: 8818–8823.
Walsh D, Zaikova E, Howes C, Song Y, Wright J, Tringe S et al (2009). Metagenome of a versatile chemolithoautotroph from expanding oceanic dead zones. Science 326: 578–582.
Weidler G, Dornmayr-Pfaffenhuemer M, Gerbl F, Heinen W, Stan-Lotter H . (2007). Communities of archaea and bacteria in a subsurface radioactive thermal spring in the Austrian Central Alps, and evidence of ammonia-oxidizing Crenarchaeota. Appl Environ Microbiol 73: 259–270.
Yutin N, Koonin EV . (2012). Archaeal origin of tubulin. Biol Direct 7: 10.
Zhang C, Ye Q, Huang Z, Li W, Chen J, Song Z et al (2008). Global occurrence of archaeal amoA genes in terrestrial hot springs. Appl Environ Microbiol 74: 6417–6426.
Acknowledgements
We appreciate support from the NSF-Integrative Graduate Education and Research Training Program to JPB and ZJJ (DGE 0654336), DOE-Pacific Northwest National Laboratory-Foundational Science Focus Area in Biological Interactions (Subcontract 112443), Douglas B Rusch at Indiana University, Bloomington, IN for NWF-PCA support, Susannah G Tringe at the DOE-JGI (Community Sequencing Program, CSP-787081) for metagenome sequencing performed at Dragon Spring (45 Mb Sanger) and Beowulf Spring (60 Mb 454), the J Craig Venter Institute for early Sanger sequencing at Beowulf Spring (12 Mb Sanger) and Justin O’Dea for positive amoA gene controls. The authors also appreciate comments from three anonymous reviewers that improved the quality of the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Beam, J., Jay, Z., Kozubal, M. et al. Niche specialization of novel Thaumarchaeota to oxic and hypoxic acidic geothermal springs of Yellowstone National Park. ISME J 8, 938–951 (2014). https://doi.org/10.1038/ismej.2013.193
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2013.193
Keywords
This article is cited by
-
Metagenomic characterization of a novel non-ammonia-oxidizing Thaumarchaeota from hadal sediment
Microbiome (2024)
-
Genome-resolved metagenomics inferred novel insights into the microbial community, metabolic pathways, and biomining potential of Malanjkhand acidic copper mine tailings
Environmental Science and Pollution Research (2023)
-
Conserved and lineage-specific hypothetical proteins may have played a central role in the rise and diversification of major archaeal groups
BMC Biology (2022)
-
Diverse ecophysiological adaptations of subsurface Thaumarchaeota in floodplain sediments revealed through genome-resolved metagenomics
The ISME Journal (2022)
-
DDX1 from Cherry valley duck mediates signaling pathways and anti-NDRV activity
Veterinary Research (2021)