Abstract
We compared the function and composition of free-living and particle-associated microbial communities at an inshore site in coastal North Carolina and across a depth profile on the Blake Ridge (offshore). Hydrolysis rates of six different polysaccharide substrates were compared for particle-associated (>3 μm) and free-living (<3 to 0.2 μm) microbial communities. The 16S rRNA- and rDNA-based clone libraries were produced from the same filters used to measure hydrolysis rates. Particle-associated and free-living communities resembled one another; they also showed similar enzymatic hydrolysis rates and substrate preferences. All six polysaccharides were hydrolyzed inshore. Offshore, only a subset was hydrolyzed in surface water and at depths of 146 and 505 m; just three polysaccharides were hydrolyzed at 505 m. The spectrum of bacterial taxa changed more subtly between inshore and offshore surface waters, but changed greatly with depth offshore. None of the OTUs occurred at all sites: 27 out of the 28 major OTUs defined in this study were found either exclusively in a surface or in a mid-depth/bottom water sample. This distinction was evident with both 16S rRNA and rDNA analyses. At the offshore site, despite the low community overlap, bacterial communities maintained a degree of functional redundancy on the whole bacterial community level with respect to hydrolysis of high-molecular-weight substrates.
Similar content being viewed by others
Introduction
Marine bacteria determine the pathways and pace of numerous biogeochemical cycles. Heterotrophic microbial communities, in particular, are responsible for remineralizing and transforming a considerable fraction of autochthonous marine organic matter (Azam, 1998) that comprises most of the organic matter in marine systems (Hedges, 1992). These communities convert and respire dissolved organic matter, colonize and remineralize particles and aggregates and help shape the nature and quantity of carbon and nutrients that pass from surface waters to the deep ocean.
The first step in the microbial transformation of complex organic matter (autochthonous and terrestrial) is the hydrolysis of high-molecular-weight compounds by extracellular enzymes that yields sufficiently small substrates (ca. 600 Da) for uptake into the cell (Weiss et al., 1991). These enzymes are produced by a fraction of the heterotrophic microbial community and may also supply nonproducers with hydrolysate (Alldredge et al., 1986; Herndl, 1988; Alldredge and Gotschalk, 1990; Simon et al., 1990; Vetter and Deming, 1999). In particles and aggregates such as marine snow, enzyme activities (and overall rates of microbial metabolism) are typically higher than in the surrounding waters, and hence are considered biogeochemical hot spots (Karner and Herndl, 1992; Smith et al., 1992; Ziervogel and Arnosti, 2008; Ziervogel et al., 2010).
The nature and composition of aggregate-associated microbial communities compared with their free-living counterparts has therefore become a focal point of investigation (DeLong et al., 1993; Acinas et al., 1999; Crump et al., 1999; Moeseneder et al., 2001; Ghiglione et al., 2009). However, these studies have provided little evidence of a consistent trend in community composition based solely on whether a community is free living or particle associated. Recently, it has been found that some bacterial types may switch lifestyles, depending on chemical triggers and substrate availability (Grossart, 2010). Bacterial community composition has also been shown to vary with many other factors, such as organic matter sources, particle residence times and geographic location (Crump et al., 1999; Zhang et al., 2007; Fuhrman et al., 2008). In addition, light or nutrient gradients have been shown to influence bacterial community composition and create zonation patterns (Wright et al., 1997; Rocap et al., 2003; Carlson et al., 2008). Several large-scale investigations have recently confirmed a divide between community composition in surface waters (Pommier et al., 2007; Fuhrman et al., 2008; Wietz et al., 2010) and in deep waters (DeLong et al., 2006; Sogin et al., 2006; Schattenhofer et al., 2009; Agogué et al., 2011). These bacterial community shifts are not the only source of divergent activity patterns; cell-free enzymes can at times contribute substantially to substrate hydrolysis (Somville and Billen, 1983; Vetter and Deming, 1999; Pantoja and Lee, 1999; Ziervogel and Arnosti, 2008; Baltar et al., 2010).
In this study, we investigated the links among metabolic capabilities, particle association and microbial community composition in waters of coastal North Carolina and across a nearby offshore depth gradient. Previous investigations of particle-associated and free-living communities have relied on the use of small substrate proxies for the determination of enzyme activities (Karner and Herndl, 1992; Crump et al., 1998; Azúa et al., 2003; Zoppini et al., 2005). Such substrate proxies provide no information on activities of endo-acting enzymes that cleave polysaccharides mid-chain, an essential step in hydrolysis of organic matter (see Arnosti, 2011 for a review). Here, we used high-molecular-weight substrates, rather than substrate proxies, to investigate the capabilities of particle-associated microbial communities. For the first time, potential enzyme activities were measured directly from the same filters used for community composition analysis.
The potential activities of polysaccharide-hydrolyzing endo-acting enzymes were the focal point because high-molecular-weight carbohydrates constitute a large percentage of phytoplankton detritus as well as marine particulate and dissolved organic matter (Painter, 1983; Benner et al., 1992; Biersmith and Benner, 1998; Lee et al., 2004). Extracellular enzyme activities of microbial communities have been shown to vary by location and depth in the ocean (Arnosti et al., 2005; Arnosti, 2011; Steen et al., 2012) and can change through the process of aggregate formation (Ziervogel et al., 2010). Moreover, different bacterial groups differ in their potential rates and enzymatic capabilities, as shown by bacterial isolates (Martinez et al., 1996; Wegner et al., 2013), genomic investigation (Glöckner et al., 2003; Bauer et al., 2006; Weiner et al., 2008; Wegner et al., 2013; Zimmerman et al., 2013) and combined measurements of enzyme activities and microbial community composition in the field (Teske et al., 2011; Arnosti et al., 2012).
To determine the identities of the most active members of microbial communities, we constructed 16S rRNA-based clone libraries in addition to conventional 16S rRNA gene libraries. Dead or dormant organisms may still possess intact DNA (Holm-Hansen et al., 1968), but rRNA is expected to degrade quickly in the environment. In this manner, our sequencing strategy compares particle-associated and free-living microbial communities in both a coastal and an offshore setting (Crump et al., 1999; Garneau et al., 2009) in combination with rRNA-based identification of active microbial community members (Moeseneder et al., 2001, 2005; Mills et al., 2005; Ghiglione et al., 2009; La Cono et al., 2009). By obtaining nearly full-length sequences of the 16S rRNA gene for identification of the most abundant and active bacterial community members, we were able to investigate the compositional patterns of particle-associated and free-living bacterial communities, as well as their abilities to access a range of high-molecular-weight polysaccharides.
Materials and methods
Sample sites and sampling
Coastal surface water was collected at ca. 1 m water depth (total water depth ca. 3–4 m) from a pier at the University of North Carolina Institute of Marine Sciences (34°43′19.32′′ N, 76°45′7.37′′ W) on the north shore of Bogue Sound, behind the barrier islands near Cape Lookout, NC, in late October 2009. A 5-l carboy was triple rinsed and filled with surface water (20 °C and 34 PSU (practical salinity unit)) collected at the end of the pier. Offshore samples were collected in early December 2009 from the R/V Cape Hatteras at a station close to the shelf break of the Blake Ridge (34°11′02.399′′ N 75°49′22.799′′ W, water depth of 530 m), ∼80 km off Cape Lookout (Supplementary Figure S1a). Three separate 5-l carboys were triple rinsed, and each was filled with water collected from different depths: 2 m ‘surface’, 146 m ‘mid-depth’ (sub-thermocline) and 505 m ‘bottom’, using a rosette of Niskin bottles attached to a Sea Bird (Bellevue, WA, USA) conductivity-temperature-depth (CTD) profiler with an SBE43 oxygen sensor and a Chelsea/Seatech/WET Labs CStar beam transmission sensor (Supplementary Figure S1b).
Sampling and filter preparation
Water samples were gravity filtered through a 3.0 μm pore size 45 mm diameter Isopore membrane filter (Millipore Corp., Bedford, MA, USA) that was placed in a separate filter holder upstream from a 0.2 μm pore size 45 mm diameter Durapore membrane filter (Millipore Corp.) in order to separate the particle-associated from the free-living community. These filter size cutoffs were chosen following other studies that have investigated free-living and particle-attached communities (Crump et al., 1999; Smith et al., 2013). Filtration occurred at 4 °C in the dark overnight. The coastal station sample was filtered on the day of collection. Over 17 h of filtration, 1.5-l of water passed through both filters. The gravity filtration of the offshore station carboys began on shore on 14 December, 10 days after collection. The carboys were stored at 4 °C in the dark before filtration. The surface water was filtered overnight on the first day (3-l filtered), the mid-depth water was filtered on the second day (3-l filtered) and the bottom water was filtered on the third day (3-l filtered). Immediately after filtration, each filter was carefully sliced with sterile razor blades, and one quarter of each filter was frozen at −80 °C for microbial community composition analysis. The remaining three quarters of the filter were cut into 12 equal pieces for the immediate measurement of hydrolytic enzyme activities, using two pieces for each polysaccharide substrate. The microbial community collected on the filter was then immediately incubated to determine its enzymatic potential in time course assays.
Enzymatic hydrolysis measurements
To assess the potential activities and substrate specificities of different microbial extracellular enzymes in all samples, we measured the hydrolysis of six different fluorescently labeled polysaccharides by microbial communities collected on the 3 and 0.2 μm filter samples. We also measured hydrolysis in unfiltered water to assess the possible effects of filtration on hydrolysis rates and patterns.
Extracellular enzymatic activities were measured (Arnosti, 1995, 2003) using six fluorescently labeled polysaccharides: pullulan, an α(1,6)-linked polymer of maltotriose; laminarin, β(1,3) glucose; xylan, β(1,4) xylose; fucoidan, a sulfated fucose polysaccharide; arabinogalactan, a mixed polymer of galactose and arabinose; and chondroitin sulfate, a sulfated polymer of N-acetyl galactoseamine and glucuronic acid. The polysaccharides (Sigma-Aldrich, St Louis, MO, USA) were labeled with fluoresceinamine and characterized as previously described (Arnosti, 2003).
Hydrolysis rates were measured using sections of the same filters used for molecular analysis. The filter pieces were placed into separate incubation vials containing 30 ml of autoclaved artificial sea water (Sigma-Aldrich). Killed controls consisted of vials containing autoclaved filter pieces in autoclaved artificial sea water. Triplicate incubations were also made with unfiltered water and with filtrate (remaining after passage through the 3 and 0.2 μm filters) for all samples and for the autoclaved controls. Substrates were added to all incubations and controls to a final concentration of 3.5 μM monomer equivalent. The coastal, offshore surface and offshore mid-depth samples were incubated at room temperature in the dark. The offshore bottom water was incubated at 5 °C in the dark. For the coastal incubations, subsamples (each ca. 1.5 ml) of sea water or artificial sea water were collected immediately after the substrate addition and after 1, 2, 6, 8, 13 and 29 days. The offshore incubations were subsampled immediately after substrate addition and after 2, 7, 14, 21 and 28 days. The subsamples were filtered through 0.2 μm pore size filters and stored frozen (−20 °C) until analysis. Samples were measured using a gel permeation chromatography/high-performance liquid chromatography system with fluorescence detection, and polysaccharide hydrolysis rates were calculated as described previously (Arnosti, 1995, 2003). All rates should be regarded as potential rates because added substrate competes with naturally occurring substrate for enzyme active sites. The concentration of added substrate (on a carbon-equivalent basis) was approximately equal to 10% of the coastal and 20% of the offshore total dissolved organic carbon (DOC).
For each site (coastal, offshore at each of three depths), the sum of the hydrolysis rate measured for each of the six substrates is referred to as the ‘summed hydrolysis rate’. We compared the potential hydrolysis rates at day 2 and at day 7 (offshore samples) or day 8 (coastal samples), time points by which most of the activities ultimately detected had developed. Over the entire time course of the experiment (28 days coastal, 29 days offshore), maximum rates of hydrolysis for each substrate were observed at or before 13 (coastal) or 14 (offshore) days; a detailed discussion of the time course is included as Supplementary Text and Supplementary Figure S2.
Cell counts and DOC measurements
Seawater samples were fixed with formaldehyde (2% v/v final concentration) shortly after sampling and stored at 4 °C. Bacteria were counted using 6-diamidino-2-phenylindole (0.1 μg l−1 final concentration) as described previously (Porter and Feig, 1980). Using an epifluorescence microscope (Olympus, Tokyo, Japan) at × 1000 magnification equipped with a digital camera (Olympus TH4-100), cells from 10 randomly chosen frames were counted on duplicate slides. Next, 10 ml of the 0.2 μm filtered seawater samples was stored in combusted glass vials at −20 °C for DOC analysis. The samples were thawed, and an aliquot was withdrawn and acidified with phosphoric acid (50% v/v) to measure DOC concentrations by high temperature catalytic oxidation using a Shimadzu (Kyoto, Japan) TOC-5000.
Sequencing and phylogenetic analysis
Nucleic acid extraction, PCR and reverse transcriptase-PCR, cloning, sequencing and chimera checks of nearly full-length 16S rRNA and 16S rRNA gene sequences (Escherichia coli nucleotide positions 28 to 1492) were performed according to standard methods (Dempster et al., 1999; Teske et al., 2002; Thompson et al., 2002; Huber et al., 2004; see Supplementary Material for details on modifications). The sequences have Genbank accession numbers JN232982 to JN233703 and JN412852, as included in the heat map (Supplementary Figure S3). Phylogenetic trees were constructed from 38 to 55 nonchimeric sequences for each library using the SILVA Release 104 SSU Ref database from www.arb-silva.de (Pruesse et al., 2007) and the ARB Software environment, version 5.1 (Ludwig et al., 2004). Preliminary ARB neighbor-joining trees were constructed, and clones forming long branches that were not clustering with well-documented marine pelagic bacterial lineages were manually rechecked for chimeras by BLASTing both ends separately. Operational taxonomic units (OTUs) were clustered at a 97% similarity cutoff using the furthest neighbor algorithm in mothur (Schloss et al., 2009). For each aligned sequence data set, a model of best-fit nucleotide substitution was statistically determined using the jModeltest program (Posada, 2008) that uses PHYML (Guindon and Gascuel, 2003). The Akaike and Bayesian information criteria selection strategies chose the same model for each tree, which was the General Time Reversible plus Invariant sites plus Gamma (GTR+I+G) distributed model. Optimal distance trees were determined using the heuristic search mode of PAUP (Swofford, 2003) and employing the maximum likelihood parameters from jModeltest for the proportion of invariant sites, nucleotide frequencies, substitution rate matrix and gamma shape. The tree topologies were checked by bootstrap analysis (using the same parameters) based on 200 replicates. A heat map was constructed using all OTUs that represented five or more sequences to show their representation percentage within each clone library (Supplementary Figure S3). Rarefaction curves were constructed for each library using mothur (Supplementary Figure S4). The supplementary phylogenetic trees (Supplementary Figures S5a–e) also include the Genbank accession numbers. The FastUniFrac web program (Hamady et al., 2009) was used for weighted and unweighted clustering and jackknife analysis of the 16S rRNA clone libraries.
Sample storage considerations
The offshore samples were stored for 10 days before filtration in the cold and dark, conditions that may have nevertheless affected community composition, even when assuming low community turnover because of an average generation time of ca. 100 h for pelagic bacteria in open-ocean waters (Yoshinaga et al., 1991) or ca. 10 to 100 h for pelagic bacteria in coastal waters (Azam and Fuhrman, 1984; Iriberri et al., 1990). Slow gravity filtration for >10 h may also affect RNA yields. However, the consistent detection of dominant uncultured bacterioplankton groups throughout all samples on DNA and RNA level, the strong differences in community composition in response to water mass and depth sampled and the absence of rRNA response peaks of enrichment-sensitive Gammaproteobacteria (especially the sensitive bottle enrichment marker Colwellia; Stewart et al., 2012) indicate that bottle effects in the unavoidable time interval between water sampling and cell capture by filtration were minimal. However, it is possible that the experimental design of this study has affected similarity analyses between free-living and particle-associated communities.
Results and discussion
Physical, chemical and biological characterization of offshore and coastal water masses
Three different water masses distinguished by CTD profile were sampled offshore (80 km from the coast), and additional samples were obtained from one coastal site (Supplementary Figure S1a). Offshore, at the surface, the temperature is characteristic of modified Gulf Stream surface water. The salinity maximum at mid-depth is indicative of the Subtropical Underwater layer, formed when evaporation increases salinity in Sargasso Sea surface water. This layer is relatively nutrient rich (Kameo et al., 2004). Because of its higher density, this intermediate water mass sinks below the surface water and overlays fresher but colder water below (Worthington, 1982; O’Connor et al., 2005). The oxygen profile from our CTD measurements (Supplementary Figure S1b) indicates an oxygenated mixed surface layer down to 70 m depth, an oxygen minimum beginning at mid-depth (110 m) and more oxygenated bottom water (below 450 m). This profile is similar to a profile measured in a previous study at nearly the same location (Cléroux et al., 2009).
The coastal water (ca. 20 °C) was cooler than the offshore surface water (25 °C) and also less saline (ca. 34 PSU compared with 36.3 PSU). DOC concentrations offshore were similar at all depths (113±1, 81±2 and 103±37 μM DOC for the surface, mid-depth and bottom samples, respectively) and were ∼45% of DOC concentration at the coastal station (221±21 μM DOC).
Connecting bacterial communities and enzyme activity
The relationship between microbial communities and their extracellular activity is complex and dynamic. Differences in enzymatic capabilities in distinct regions of the ocean have been associated with differences in microbial community structure (Arnosti et al., 2011, 2012); conversely, changes in enzymatic requirements because of substrate availability or other environmental parameters can drive changes in the structure of bacterial communities (Grossart et al., 2007; Haynes et al., 2007; Murray et al., 2007; Grossart, 2010; Burke et al., 2011; Teeling et al., 2012). The enzymatic capabilities of microbial communities are likely controlled by a combination of specific hydrolytic capabilities of different bacterial types and feeding guilds, and their differential expression of enzymes (Arnosti, 2011). The measurement of enzymatic activities with fluorescently labeled polysaccharides integrates these responses to bulk community functions that cannot yet be assigned to individual community members or taxonomic subgroups. Because of the time course incubations necessary for the measurement of hydrolysis rates, microbial communities can respond to substrates either by the activity of enzymes already present (associated with the cell or excreted to the environment), by the induction of enzymes or by the growth of specific members of the community. The experimental design of this study links potential substrate hydrolysis rates as community functions to community composition assessed at genus- and species-level taxonomic resolution by clone libraries that provide a measure of the extent and active members of the community at initiation of the hydrolysis experiments. In the following section we discuss inshore–offshore patterns in enzyme activities and microbial communities, depth patterns, comparisons of particle-associated and free-living communities and finally conclude by discussing the links between bacterial community composition and potential enzymatic activity.
Inshore–offshore differences in enzyme activities and microbial communities
Summed hydrolysis rates in whole water after 2 days of incubation were somewhat higher in coastal than in offshore waters (Figure 1a), but these differences in summed rates did not scale with the order-of-magnitude greater number of cells at the coastal station compared with the offshore station (Supplementary Figure S6). A comparison of hydrolysis rates after 7 or 8 days of incubation likewise shows only moderate differences in summed hydrolysis rates between coastal and offshore surface samples (Figures 1c and d).
Enzymatic hydrolysis rates for coastal and offshore samples. (a) Hydrolysis rates for each substrate in whole water at 2 days; (b) hydrolysis rates for free-living (FL) and particle-associated (PA) fractions at 2 days; (c) hydrolysis rates for each substrate in whole water at 7 or 8 days; (d) hydrolysis rates for FL and PA fractions at 7 or 8 days. Bars show deviation between two replicates. See Supplementary Figure S2 for time courses of hydrolysis.
The spectrum of substrates hydrolyzed, however, differed notably between the coastal and offshore stations. At the coastal station, all six substrates were readily hydrolyzed in whole water after 2 days of incubation (Figure 1a), but only three or four of the substrates were hydrolyzed in whole water from the different depths at the offshore station. After 7 or 8 days of incubation, hydrolysis of chondroitin became evident for the surface water offshore (Figure 1c). The spectrum of substrates hydrolyzed at the offshore station, however, remained narrower than at the coastal station, even upon extended incubation (Supplementary Figure S2). In the particle-associated and free-living fractions, initially a narrower spectrum of substrates was hydrolyzed (Figure 1b) compared with the whole water (Figure 1a), but with extended incubation time (Figure 1d; see also Supplementary Figure S2), the spectrum of substrates hydrolyzed in each sample became the same as initially observed for the corresponding location/depth in the whole water sample: six substrates for the coastal sample, and three or four substrates for the offshore station (compare Figures 1a and d; note that pullulan hydrolysis was first detected in the mid-depth offshore samples after days 7/8; see Supplementary Figure S2).
The observation that somewhat extended incubation times were necessary to detect in the filter samples the range of activities initially observed in the whole water may be because of an initial reduction in cell numbers/activity immediately after filtration. As discussed at length in Arnosti (2003), detection of specific enzyme activities using fluorescently labeled polysaccharides requires hydrolysis of a sufficient portion of the total added polysaccharide to a molecular weight sufficiently small for the change to be resolved by gel permeation chromatography. A smaller starting population or a temporary reduction in activity of prokaryotes thus requires somewhat more time for hydrolysis to be detectable. This idea is supported by the observation that the spectrum of substrates ultimately hydrolyzed was the same for a sample from a given location (for example, for offshore surface water) for the whole water as for the filter-fractionated water samples (FL+PA), and that the maximum rate observed for whole water and for the filter-fractionated water samples (FL+PA) were also similar (compare the sum of the FL+PA rates in Figure 1d with the rates for whole water in Figure 1c).
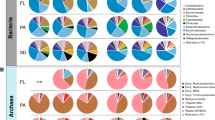
Similar to the patterns of enzyme activities, patterns of 16S rDNA and rRNA sequences showed spatial distinctions. In general, the 16S rDNA and 16S rRNA sequences were commonly affiliated with specific bacterial lineages characteristic for the marine pelagic environment. Full-length phylotypes in this survey could be reliably identified at genus or intragenus level when culture isolates were available. Most conspicuously, all but one of the coastal and offshore libraries from surface waters included a high proportion of Alphaproteobacteria (Figure 2 and Supplementary Figure S5a); the exception was the nearshore, particulate, DNA-based library (discussed below). The mid-depth and bottom libraries from colder, deeper water offshore were dominated by Gammaproteobacteria (Figure 2 and Supplementary Figure S5b). This difference is consistent with patterns reported from other locations in the North Atlantic (Sogin et al., 2006; Lauro and Bartlett, 2008; Schattenhofer et al., 2009; Agogué et al., 2011). The offshore and coastal surface station libraries were similar and shared the SAR11 cluster, Roseobacter, Bacteroidetes and photosynthetic groups (Figure 2 and Supplementary Figure S5c), although Actinobacteria were found in the coastal samples but not offshore. Generally, these bacterial groups have been found in global surveys of ocean surface waters (Weitz et al., 2010), and many of the coastal station surface groups match those found previously off Cape Hatteras (Rappé et al., 1997). Whereas the rDNA and rRNA clone libraries from coastal surface waters were dominated by SAR11 and Roseobacter clones, respectively, other groups predominated in some of the offshore surface libraries (Figure 2). The particle-associated DNA library contained 33% clones closely related to the genus Pseudoalteromonas within the Gammaproteobacteria; the free-living RNA library contained 51% clones close to the genus Thalassospira within the Alphaproteobacteria.
Bacterial community composition for all 16S rRNA and rRNA gene clone libraries. Taxonomic specificity ranges from phylum- to genus-level resolution when applicable. Alphaproteobacteria groups are shown in blue. Gammaproteobacteria groups are shown in red. Actino., Actinobacteria; Cyano., Cyanobacteria; Flavo., Flavobacteria; Pseudoaltero., Pseudoalteromonas. See Supplementary Information for color codes and specific OTUs.
The clone libraries showed distinct differences with depth. The mid-depth libraries from the Subtropical Underwater (Supplementary Figure S1b) showed distinctly low diversity; their unusual bacterial communities were dominated by Gammaproteobacteria, especially phylotypes related to the genera Marinomonas, Oceaniserpentilla and Balneatrix (Figure 2). Sequences affiliated with these genera occurred consistently in all four (particle associated and free living, and DNA and RNA derived) mid-depth libraries; these groups were otherwise found in lower relative abundance in the offshore station bottom libraries. A cluster of clones related to the genus Oceaniserpentilla (Supplementary Figure S5b) was exclusive to the mid-depth libraries. Close relatives (99.6% nucleotide similarity) of this group, termed the Deepwater Horizon Oceanospirillales, were found to dominate water samples polluted by the subsurface hydrocarbon plume caused by the Deepwater Horizon oil spill in the Gulf of Mexico in 2010 (Hazen et al., 2010; Mason et al., 2012). Within the widely occurring Bacteroidetes phylum, an uncultured group originally found in the Atlantic Ocean (Alonso-Sáez et al., 2006) dominated the mid-depth layer, and North Sea Flavobacterial groups (Alonso et al., 2007) dominated the coastal and offshore surface libraries (Supplementary Figure S5c).
In the offshore station bottom libraries, members of the genus Colwellia and the SUP05 cluster within the Gammaproteobacteria were observed (Walsh et al., 2009; Marshall and Morris, 2012), in addition to Bacteroidetes, Actinobacteria and Deltaproteobacteria (mostly SAR324; Wright et al., 1997). Based on single-cell genome sequencing, SAR324 (Supplementary Figure S5d) and SUP05 (Supplementary Figure S5b) were recently identified as candidate autotrophs (Swan et al., 2011). The bottom free-living RNA library was distinct from all of the other clone libraries. This library was dominated by Actinobacteria and Deltaproteobacteria (Figure 2 and Supplementary Figure S5e) and mostly lacked the Gammaproteobacteria that dominated the other deep and mid-water clone libraries in this survey.
Depth effects
Although bacterial communities changed with depth extensively from genus- to phylum-level composition (see Supplementary Text for details), the ability to hydrolyze a particular substrate or set of substrates with depth changed more gradually (Figure 1). A narrower spectrum (fewer substrates) were hydrolyzed at the deepest depth for all samples—particle associated, free living and unfiltered water, with the bottom water communities hydrolyzing only laminarin, xylan and chondroitin, but not pullulan (or fucoidan or arabinogalactan that were hydrolyzed at the coastal station; Figure 1).
Greater selectivity in substrate hydrolysis with depth did not scale with changes in rates of hydrolysis of specific substrates, or with cell numbers. Laminarin, chondroitin and xylan hydrolysis rates, for example, did not change systematically with depth offshore (Figure 1). Moreover, summed hydrolysis rates on a per-cell basis in the mid-depth and bottom waters were approximately twice as high as in surface waters (Supplementary Figure S6). Both of these results—a disconnect between cell numbers and summed activities, and increased (calculated) per-cell enzyme activities with depth—have been observed previously (Arnosti et al., 2009; Baltar et al., 2009). For example, cell counts in the mesopelagic ocean decreased faster with depth than hydrolytic activities (measured with small substrate proxies), such that the calculated per-cell activity increased with depth; these data implied a greater enzymatic production and demand for cellular carbon at depth (Baltar et al., 2009). We find the same trend of increased extracellular enzyme activity per cell at depth, even with the loss of the ability to hydrolyze certain substrates (Figure 1). However, there was no significant reduction in summed hydrolysis rates with depth, and this is unusual compared with results from the few other investigations of polysaccharide hydrolysis over depth profiles (Arnosti et al., 2005; Steen et al., 2008, 2012). Ultimately, hydrolysis rates are a function of enzyme quantity and turnover number, parameters that cannot be defined in environmental samples with currently available techniques.
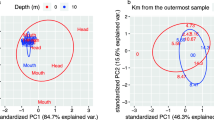
Depth-related changes in microbial community composition were quite distinct. A cluster diagram (Figure 3a) and heat map (Supplementary Figure S3) illustrate strong partitioning between the surface water and the deepwater column communities, often on the level of specific pelagic lineages that are defined by representative 16S rRNA gene phylotypes. With the exception of phylotypes within the genus Pseudomonas, all major OTUs (with more than five sequences) were found exclusively in either a surface-water station library (coastal or surface offshore) or in an offshore mid-depth or bottom site library (Supplementary Figure S3). The six most commonly found OTUs, which represented 37% of all clones, partitioned in this manner. None of the OTUs was found in all of the samples; none of the OTUs was found at all depths in the offshore station. A dendrogram created with Fast UniFrac analysis robustly confirms the surface–subsurface divide; whether OTU abundances are taken into account (Figure 3a) or not (Supplementary Figure S7), most surface-water libraries form a group distinct from the subsurface water column libraries. Dendograms of enzyme activities after 2 days and after 7/8 days of incubation illustrate the evolution in hydrolysis patterns with time, with increasing separation of the coastal particle-associated fraction from the other fractions (Figures 3b and c). The coastal particle-associated fraction developed the full spectrum of 6 activities at 7/8 days, whereas the free-living coastal fraction only showed all activities at the 13 day time point (Supplementary Figure S2).
(a) Cluster diagram of all 16S rDNA and rRNA clone libraries, constructed with fast UniFrac using the weighted option that takes into account OTU abundances. The scale bar shows the distance between clusters in UniFrac units: a distance of 0 means that two samples are identical, and a distance of 0.5 means that two samples contain mutually exclusive lineages. Particle-associated libraries are in bold. The numbers show the support for each node as determined by jackknife analysis. FL, free living; PA, particle associated. (b) Cluster diagram of enzyme activities after incubation for 2 days. (c) Cluster diagram of enzyme activities after incubation for 7 or 8 days. See Figure 1 for hydrolysis rates. Pairwise Euclidean distances between the samples were calculated using the software program R. Bootstrap numbers shown are the approximately unbiased P-values that were calculated using the R package pvclust (Suzuki and Shimodaira, 2011) with the Ward method.
In the mid-depth offshore libraries, all bacterial lineages represented in the DNA clone libraries were also represented in the RNA libraries (but not vice versa) (Supplementary Figure S3). In other words, bacterial cells that contribute substantially to the DNA pool are also active, possibly indicating tight coupling between bacterial community composition and activity. This trend toward tighter coupling of bacterial presence and activity was previously attributed to decreased oxidative stress in anoxic deep water (Brettar et al., 2012) but is documented here in oxygenated deep waters. In contrast to the mid-water sample, the offshore surface and deepwater samples as well as the coastal station contain several OTUs that appear on DNA level but without counterparts in the RNA data sets, indicating some bacterial populations with attenuated activity and rRNA content, or entirely inactive populations (Supplementary Figure S3).
Particle-associated and free-living community composition and hydrolytic activity
The extent to which particle-attached and free-living communities differ likely depends in part on particle integrity and residence time in a given location. It has been suggested that longer particle residence times, which are often found in estuaries and coastal systems, favor the development of phylogenetically distinct particle-associated and free-living groups (Crump et al., 1999, 2004; Selje and Simon, 2003; Zhang et al., 2007), but this trend is not observed in all coastal areas (Crump et al., 1999; Garneau et al., 2009). In contrast, Acinas et al. (1999) saw stronger differences between particle-associated and free-living bacteria at the surface than at depth (400 m) in the Mediterranean. Moeseneder et al. (2001) saw consistent differences between particle-associated and free-living bacterial communities at all depths (down to 1000 m) in the Aegean Sea, where temperature and salinity gradients were minor or nonexistent with depth. In this data set, the compositional differences between the particle-associated and the free-living communities did not change systematically with depth (Figure 2). Particle-associated and free-living communities exhibited similar summed hydrolysis rates (Figures 1b and d), although the time point at which some enzyme activities were detected differed (Supplementary Figure S2). In any case, at each depth the particle-associated and free-living water fractions ultimately hydrolyzed the same spectrum of substrates as the unfiltered fraction (Figure 1 and Supplementary Figure S2). In the filtrate (<0.2 μm), hydrolysis was only measurable for laminarin, and only in the coastal station sample (data not shown), suggesting little contribution from freely dissolved enzymes to substrate hydrolysis at these locations. Similar contributions by particle-associated and free-living bacteria to the degradation of organic matter have been observed in other studies, including in the degradation of Phaeocystis biomass (Becquevort et al., 1998) and in rates of hydrolysis of bacterial isolates from the Southern California Bight (Martinez et al., 1996). However, in some circumstances, hydrolytic activity has varied between particle-associated and free-living bacteria (Azúa et al., 2003).
Functional redundancy in enzymatic capabilities
Defining the complex linkages between microbial community composition and hydrolytic capabilities is important in order to understand the carbon cycling capabilities of microbial communities in the ocean. Pelagic microbial communities from diverse locations can differ considerably in hydrolytic capabilities (see, for example, Arnosti et al., 2005, 2011, 2012; Steen et al., 2012); yet, two compositionally different communities in surface and bottom waters of an Arctic fjord demonstrated similar hydrolytic capabilities and thus functional redundancy (Teske et al., 2011). Functional redundancy in enzymatic capabilities occurs when multiple groups of bacteria share the capacity to use a substrate, often a substrate that is widely available. As an example, laminarin (a common algal storage product; Painter, 1983) was hydrolyzed quickly by all samples in this study and others (Arnosti, 2008; Steen et al., 2008; Arnosti et al., 2009), possibly because of the wide distribution of laminarin hydrolytic capability across microbial phyla and subphyla. Several cultured marine bacteria (Pseudoalteromonas tetraodonis, Sulfitobacter pontiacus and Vibrio splendidus) that are closely related to Gamma- and Alpha-proteobacterial phylotypes in our clone libraries have been shown to grow on laminarin as a sole carbon source (Alderkamp et al., 2007). In contrast, fucoidan hydrolysis is often minimal or not measurable in the water column (Arnosti, 2004; Arnosti et al., 2005, 2011; Teske et al., 2011), and fucose-containing polysaccharides have been identified as remineralization ‘problems’ (Amon et al., 2001) even though ubiquitous transparent exopolymers contain high concentrations of sulfated deoxysugars with composition corresponding to fucoidan (Alldredge et al., 1993; Mopper et al., 1995). The variable extent of functional redundancy among microbial communities thus complicates efforts to link specific bacteria with enzymatic function.
This study, the first to use high-molecular-weight substrates to compare hydrolytic activity and community composition from the same filters, demonstrates several biogeographic trends of bacterial diversity and function in the marine water column:
-
a)
Partitioning of microbial community composition was apparent in (1) the contrast between surface waters (coastal and offshore) and offshore subsurface waters; and (2) some differences among the three offshore water depths, where vertical particle flux is evidently insufficient to homogenize the particle-associated communities across water depths.
-
b)
The differences in the spectrum of enzyme activities between the coastal and offshore surface stations, as well as between the surface water and bottom water offshore stations, demonstrate that the extent to which a high-molecular-weight substrate is bioavailable depends on microbial community capabilities, and not just on the chemical structure of the substrate. The similarity of summed hydrolysis rates, despite considerable differences in cell abundance, suggests that mid-depth and deep water communities maintain the ability to react strongly to the presence of substrates for which they have appropriate enzymes.
-
c)
Functional redundancy is also evident in the range of high-molecular-weight substrates hydrolyzed: regardless of the low community overlap at the genus level, bacterial communities at different water depths are apparently capable of hydrolyzing a similar set of substrates, in this case laminarin, chondroitin and xylan. Apparently, representatives of many microbial lineages may have the correct enzymes to access specific polysaccharides.
-
d)
Nonetheless, community differences with depth are consistent with the loss of certain enzymatic activities in deep water compared with surface waters (Figure 1), suggesting fundamental differences in the abilities of these communities to access diverse substrates. The changing taxonomic profiles of the bacterial community in the water column ultimately translate into changing hydrolysis patterns and show the range as well as the limits of functional redundancy.
References
Acinas S, Anton J, Rodriguez-Valera F . (1999). Diversity of free-living and attached bacteria in offshore western Mediterranean waters as depicted by analysis of genes encoding 16S rRNA. Appl Environ Microbiol 65: 514–522.
Agogué H, Lamy D, Neal PR, Sogin ML, Herndl GJ . (2011). Water mass–specificity of bacterial communities in the North Atlantic revealed by massively parallel sequencing. Mol Ecol 20: 258–274.
Alderkamp AC, Van Rijssel M, Bolhuis H . (2007). Characterization of marine bacteria and the activity of their enzyme systems involved in degradation of the algal storage glucan laminarin. FEMS Microbiol Ecol 59: 108–117.
Alldredge AL, Cole JJ, Caron DA . (1986). Production of heterotrophic bacteria inhabiting macroscopic organic aggregates (marine snow) from surface waters. Limnol Oceanogr 31: 68–78.
Alldredge AL, Gotschalk CC . (1990). The relative contribution of marine snow of different origins to biological processes in coastal waters. Cont Shelf Res 10: 41–58.
Alldredge AL, Passow U, Logan BE . (1993). The abundance and significance of a class of large, transparent organic particles in the ocean. Deep Sea Res I 40: 1131–1140.
Alonso C, Warnecke F, Amann R, Pernthaler J . (2007). High local and global diversity of Flavobacteria in marine plankton. Environ Microbiol 9: 1253–1266.
Alonso-Sáez L, Arístegui J, Pinhassi J, Gómez-Consarnau L, González JM, Vaqué D et al. (2006). Bacterial assemblage structure and carbon metabolism along a productivity gradient in the NE Atlantic Ocean. Aquat Microb Ecol 46: 43–53.
Amon RMW, Fitznar HP, Benner R . (2001). Linkages among the bioreactivity, chemical composition, and diagenetic state of marine dissolved organic matter. Limnol Oceanogr 46: 287–297.
Arnosti C . (1995). Measurement of depth- and site-related differences in polysaccharide hydrolysis rates in marine sediments. Geochim Cosmochim Acta 59: 4247–4257.
Arnosti C . (2003). Fluorescent derivatization of polysaccharides and carbohydrate-containing biopolymers for measurement of enzyme activities in complex media. J Chromatogr B 793: 181–191.
Arnosti C . (2004). Speed bumps and barricades in the carbon cycle: substrate structural effects on carbon cycling. Mar Chem 92: 263–273.
Arnosti C . (2008). Functional differences between Arctic seawater and sedimentary microbial communities: contrasts in microbial hydrolysis of complex substrates. FEMS Microbiol Ecol 66: 343–351.
Arnosti C . (2011). Microbial extracellular enzymes and the marine carbon cycle. Ann Rev Mar Sci 3: 401–425.
Arnosti C, Durkin S, Jeffrey WH . (2005). Patterns of extracellular enzyme activities among pelagic marine microbial communities: implications for cycling of dissolved organic carbon. Aquat Microb Ecol 38: 135–145.
Arnosti C, Ziervogel K, Ocampo L, Ghobrial S . (2009). Enzyme activities in the water column and in shallow permeable sediments from the northeastern Gulf of Mexico. Estuar Coast Shelf Sci 84: 202–208.
Arnosti C, Steen AD, Ziervogel K, Ghobrial S, Jeffrey WH . (2011). Latitudinal gradients in degradation of marine dissolved organic carbon. PLoS One 6: e28900.
Arnosti C, Fuchs BM, Amann R, Passow U . (2012). Contrasting extracellular enzyme activities of particle associated bacteria from distinct provinces of the North Atlantic Ocean. Front Microbiol 3: 425.
Azam F . (1998). Microbial control of oceanic carbon flux: the plot thickens. Science 280: 694–695.
Azam F, Fuhrman JA . (1984). Measurement of bacterioplankton growth in the sea and its regulation by environmental conditions. In: Hobbie JE, Williams PJ, (eds) Heterotrophic Activity in the Sea. Plenum Publishing Corp: New York, pp 179–196.
Azúa I, Unanue M, Ayo B, Artolozaga I, Arrieta JM, Iriberri J . (2003). Influence of organic matter quality in the cleavage of polymers by marine bacterial communities. J Plankton Res 25: 1451–1460.
Baltar F, Arístegui J, Sintes E, Van Aken HM, Gasol JM, Herndl GJ . (2009). Prokaryotic extracellular enzymatic activity in relation to biomass production and respiration in the meso-and bathypelagic waters of the (sub) tropical Atlantic. Environ Microbiol 11: 1998–2014.
Baltar F, Arístegui J, Gasol JM, Sintes E, Van Aken HM, Herndl GJ . (2010). High dissolved extracellular enzymatic activity in the deep central Atlantic Ocean. Aquat Microb Ecol 58: 287–302.
Bauer M, Kube M, Teeling H, Richter M, Lombardot T, Allers E et al. (2006). Whole genome analysis of the marine Bacteroidetes “Gramella forsetii” reveals adaptations to degradation of polymeric organic matter. Environ Microbiol 8: 2201–2213.
Becquevort S, Rousseau V, Lancelot C . (1998). Major and comparable roles for free-living and attached bacteria in the degradation of Phaeocystis-derived organic matter in Belgian coastal waters of the North Sea. Aquat Microb Ecol 14: 39–48.
Benner R, Pakulski J, McCarthy M, Hedges JI, Hatcher PG . (1992). Bulk chemical characteristics of dissolved organic matter in the ocean. Science 255: 1561.
Biersmith A, Benner R . (1998). Carbohydrates in phytoplankton and freshly produced dissolved organic matter. Mar Chem 63: 131–144.
Brettar I, Christen R, Hofle MG . (2012). Analysis of bacterial core communities in the central Baltic by comparative RNA-DNA-based fingerprinting provides links to structure-function relationships. ISME J 6: 195–212.
Burke C, Steinberg P, Rusch DB, Kjelleberg S, Thomas T . (2011). Bacterial community assembly based on functional genes rather than species. Proc Natl Acad Sci USA 108: 14288–14293.
Carlson CA, Morris R, Parsons R, Treusch AH, Giovannoni SJ, Vergin K . (2008). Seasonal dynamics of SAR11 populations in the euphotic and mesopelagic zones of the northwestern Sargasso Sea. ISME J 3: 283–295.
Cléroux C, Lynch-Stieglitz J, Schmidt MW, Cortijo E, Duplessy J-C . (2009). Evidence for calcification depth change of Globorotalia truncatulinoides between deglaciation and Holocene in the Western Atlantic Ocean. Mar Micropaleontol 73: 57–61.
Crump BC, Baross JA, Simenstad CA . (1998). Dominance of particle-attached bacteria in the Columbia River estuary, USA. Aquat Microb Ecol 14: 7–18.
Crump B, Armbrust E, Baross J . (1999). Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia river, its estuary, and the adjacent coastal ocean. Appl Environ Microbiol 65: 3192–3204.
Crump BC, Hopkinson CS, Sogin ML, Hobbie JE . (2004). Microbial biogeography along an estuarine salinity gradient: combined influences of bacterial growth and residence time. Appl Environ Microbiol 70: 1494–1505.
DeLong EF, Franks DG, Alldredge AL . (1993). Phylogenetic diversity of aggregate-attached vs free-living marine bacterial assemblages. Limnol Oceanogr 38: 924–934.
DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU et al. (2006). Community genomics among stratified microbial assemblages in the ocean’s interior. Science 311: 496–503.
Dempster EL, Pryor KV, Francis D, Young JE, Rogers HJ . (1999). Rapid DNA extraction from ferns for PCR-based techniques. Biotechniques 27: 66–68.
Fuhrman JA, Steele JA, Hewson I, Schwalbach MS, Brown MV, Green JL et al. (2008). A latitudinal diversity gradient in planktonic marine bacteria. Proc Natl Acad Sci USA 105: 7774–7778.
Garneau MČ, Vincent WF, Terrado R, Lovejoy C . (2009). Importance of particle-associated bacterial heterotrophy in a coastal Arctic ecosystem. J Marine Syst 75: 185–197.
Ghiglione JF, Conan P, Pujo-Pay M . (2009). Diversity of total and active free-living vs particle-attached bacteria in the euphotic zone of the NW Mediterranean Sea. FEMS Microbiol Lett 299: 9–21.
Glöckner FO, Kube M, Bauer M, Teeling H, Lombardot T, Ludwig W et al. (2003). Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc Natl Acad Sci USA 100: 8298.
Grossart H-P . (2010). Ecological consequences of bacterioplankton lifestyles: changes in concepts are needed. Environ Microb Reports 2: 706–714.
Grossart H-P, Engel A, Arnosti C, De La Rocha C, Murray A, Passow U . (2007). Microbial dynamics in autotrophic and heterotrophic seawater mesocosms: III Organic matter fluxes. Aquat Microb Ecol 49: 143–156.
Guindon S, Gascuel O . (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704.
Hamady M, Lozupone C, Knight R . (2009). Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J 4: 17–27.
Haynes K, Hofmann TA, Smith CJ, Ball AS, Underwood GJC, Osborn AM . (2007). Diatom-derived carbohydrates as factors affecting bacterial community composition in estuarine sediments. Appl Environ Microbiol 73: 6112–6124.
Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, Singh N et al. (2010). Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330: 204–208.
Hedges JI . (1992). Global biogeochemical cycles: progress and problems. Mar Chem 39: 67–93.
Herndl GJ . (1988). Ecology of amorphous aggregations (marine snow) in the Northern Adriatic Sea. II. Microbial density and activity in marine snow and its implication to overall pelagic processes. Mar Ecol Prog Ser 48: 265–275.
Holm-Hansen O, Sutcliffe WH Jr, Sharp J . (1968). Measurement of deoxyribonucleic acid in the ocean and its ecological significance. Limnol Oceanogr 13: 507–514.
Huber T, Faulkner G, Hugenholtz P . (2004). Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20: 2317–2319.
Iriberri J, Unanue M, Ayo B, Barcina I, Egea L . (1990). Bacterial production and growth rate estimation from [3H]thymidine incorporation for attached and free-living bacteria in aquatic systems. Appl Environ Microbiol 56: 483–487.
Kameo K, Shearer MC, Droxler AW, Mita I, Watanabe R, Sato T . (2004). Glacial-interglacial surface water variations in the Caribbean Sea during the last 300 ky based on calcareous nannofossil analysis. Palaeogeogr Palaeoclimatol Palaeoecol 212: 65–76.
Karner M, Herndl G . (1992). Extracellular enzymatic activity and secondary production in free-living and marine snow-associated bacteria. Marine Biol 113: 341–347.
La Cono V, Tamburini C, Genovese L, Spada GL, Denaro R, Yakimov MM . (2009). Cultivation-independent assessment of the bathypelagic archaeal diversity of Tyrrhenian Sea: Comparative study of rDNA and rRNA-derived libraries and influence of sample decompression. Deep Sea Res II 56: 768–773.
Lauro FM, Bartlett DH . (2008). Prokaryotic lifestyles in deep sea habitats. Extremophiles 12: 15–25.
Lee C, Wakeham SG, Arnosti C . (2004). Particulate organic matter in the sea: the composition conundrum. Ambio 33: 559–568.
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar et al. (2004). ARB: a software environment for sequence data. Nucl Acids Res 32: 1363–1371.
Martinez J, Smith D, Steward G, Azam F . (1996). Variability in ectohydrolytic enzyme activities of pelagic marine bacteria and its significance for substrate processing in the sea. Aquat Microb Ecol 10: 223–230.
Marshall KT, Morris RM . (2012). Isolation of an aerobic sulfur oxidizer from the SUP05/Arctic96BD-19 clade. ISME J 7: 452–455.
Mason OU, Hazen TC, Borglin S, Chain PS, Dubinsky EA, Fortney JL et al. (2012). Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to Deepwater Horizon oil spill. ISME J 6: 1715–1727.
Mills HJ, Martinez RJ, Story S, Sobecky PA . (2005). Characterization of microbial community structure in Gulf of Mexico gas hydrates: comparative analysis of DNA- and RNA-derived clone libraries. Appl Environ Microbiol 71: 3235–3247.
Moeseneder M, Winter C, Herndl G . (2001). Horizontal and vertical complexity of attached and free-living bacteria of the eastern Mediterranean Sea, determined by 16S rDNA and 16S rRNA fingerprints. Limnol Oceanogr 46: 95–107.
Moeseneder MM, Arrieta JM, Herndl GJ . (2005). A comparison of DNA-and RNA-based clone libraries from the same marine bacterioplankton community. FEMS Microbiol Ecol 51: 341–352.
Mopper K, Zhou J, Sri Ramana K, Passow U, Dam HG, Drapeau DT . (1995). The role of surface-active carbohydrates in the flocculation of a diatom bloom in a mesocosm. Deep Sea Res II 42: 47–73.
Murray AE, Arnosti C, De La Rocha CL, Grossart HP, Passow U . (2007). Microbial dynamics in autotrophic and heterotrophic seawater mesocosms. II. Bacterioplankton community structure and hydrolytic enzyme activities. Aquat Microb Ecol 49: 123–141.
O’Connor BM, Fine RA, Olson DB . (2005). A global comparison of subtropical underwater formation rates. Deep Sea Res I 52: 1569–1590.
Painter TJ Algal polysaccharides. (1983). In: Aspinall GO, (ed) The Polysaccharides vol. 2. Academic Press: New York, pp 195–285.
Pantoja S, Lee C . (1999). Peptide decomposition by extracellular hydrolysis in coastal seawater and salt marsh sediment. Mar Chem 63: 273–291.
Pommier T, Canbäck B, Riemann L, Boström KH, Simu K, Lundberg P et al. (2007). Global patterns of diversity and community structure in marine bacterioplankton. Mol Ecol 16: 867–880.
Porter KG, Feig YS . (1980). The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr 25: 943–948.
Posada D . (2008). jModelTest: Phylogenetic Model Averaging. Mol Biol Evol 25: 1253–1256.
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucl Acids Res 35: 7188–7196.
R Core Team. (2014) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria, http://www.R-project.org/.
Rappé MS, Kemp PF, Giovannoni SJ . (1997). Phylogenetic diversity of marine coastal picoplankton 16S rRNA genes cloned from the continental shelf off Cape Hatteras, North Carolina. Limnol Oceanogr 42: 811–826.
Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA et al. (2003). Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424: 1042–1047.
Schattenhofer M, Fuchs BM, Amann R, Zubkov MV, Tarran GA, Pernthaler J . (2009). Latitudinal distribution of prokaryotic picoplankton populations in the Atlantic Ocean. Environ Microbiol 11: 2078–2093.
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541.
Selje N, Simon M . (2003). Composition and dynamics of particle-associated and free-living bacterial communities in the Weser estuary, Germany. Aquat Microb Ecol 30: 221–237.
Simon M, Alldredge AL, Azam F . (1990). Bacterial carbon dynamics on marine snow. Mar Ecol Prog Ser 65: 205–211.
Smith DC, Simon M, Alldredge A, Azam F . (1992). Intense hydrolytic enzyme activity on marine aggregates and implications for rapid particle dissolution. Nature 359: 139–142.
Smith MW, Allen LZ, Allen AE, Herfort L, Simon HM . (2013). Contrasting genomic properties of free-living and particle-attached microbial assemblages within a coastal ecosystem. Front Microbiol 4: 120.
Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR et al. (2006). Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci USA 103: 12115–12120.
Somville M, Billen G . (1983). A method for determining exoproteolytic activity in natural waters. Limnol Oceangr 28: 109–193.
Steen AD, Hamdan LJ, Arnosti C . (2008). Dynamics of dissolved carbohydrates in the Chesapeake Bay: insights from enzyme activities, concentrations, and microbial metabolism. Limnol Oceanogr 53: 936–947.
Steen AD, Ziervogel K, Ghobrial S, Arnosti C . (2012). Functional variation among polysaccharide-hydrolyzing microbial communities in the Gulf of Mexico. Marine Chem 138–139: 13–20.
Stewart F, Dalsgaard T, Young CR, Thamdrup B, Revsbech NP, Ulloa O, Canfield DE, DeLong EF . (2012). Experimental incubations elicit profound changes in community transcription in OMZ bacterioplankton. PLoS One 7 (5): e37118.
Swan BK, Martinez-Garcia M, Preston CM, Sczyrba A, Woyke T, Lamy D et al. (2011). Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science 333: 1296–1300.
Suzuki R, Shimodaira H . (2011). pvclust: Hierarchical Clustering with P-Values via Multiscale Bootstrap Resampling. R package version 1.2-2. http://CRAN.R-project.org/package=pvclust.
Swofford DL . (2003) PAUP*. Phylogenetic Analysis Using Parsimony and Other Methods. Version 4. Sinauer Associates: Sunderland, MA.
Teske A, Durbin A, Ziervogel K, Cox C, Arnosti C . (2011). Microbial community composition and function in permanently cold seawater and sediments from an arctic fjord of Svalbard. Appl Environ Microbiol 77: 208–218.
Teske A, Hinrichs KU, Edgcomb V, de Vera Gomez A, Kysela D, Sylva SP et al. (2002). Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl Environ Microbiol 68: 1994–2008.
Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336: 608–611.
Thompson JR, Marcelino LA, Polz MF . (2002). Heteroduplexes in mixed-template amplifications: formation, consequence and elimination by ‘reconditioning PCR’. Nucleic Acids Res 30: 2083–2088.
Vetter Y, Deming J . (1999). Growth rates of marine bacterial isolates on particulate organic substrates solubilized by freely released extracellular enzymes. Microb Ecol 37: 86–94.
Walsh DA, Zaikova E, Howes CG, Song YC, Wright JJ, Tringe SG et al. (2009). Metagenome of a versatile chemolithoautotroph from expanding oceanic dead zones. Science 326: 578–582.
Wegner C-E, Richter-Heitmann T, Klindworth A, Klockow C, Richter M, Achstetter T et al. (2013). Expression of sulfatases in Rhodopirellula baltica and the diversity of sulfatases in the genus Rhodopirellula. Mar Genomics 9: 51–61.
Weiner RM, Taylor LE, Henrissat B, Hauser L, Land M, Coutinho PM et al. (2008). Complete genome sequence of the complex carbohydrate-degrading marine bacterium, Saccharophagus degradans Strain 2-40T. PLoS Genet 4: e1000087.
Weiss MS, Abele U, Weckesser J, Welte W, Schiltz E, Schulz GE . (1991). Molecular architecture and electrostatic properties of a bacterial porin. Science 254: 1627–1630.
Wietz M, Gram L, Jorgensen B, Schramm A . (2010). Latitudinal patterns in the abundance of major marine bacterioplankton groups. Aquat Microb Ecol 61: 179–189.
Worthington LV . (1982). North Atlantic circulation and water mass formation. J Mar Res 40: xii–xxii.
Worthington LV . (1976) On the North Atlantic circulation. John Hopkins Oceanographic Studies vol. VI. The John Hopkins University Press: Baltimore and London, p 110.
Wright T, Vergin K, Boyd P, Giovannoni S . (1997). A novel delta-subdivision proteobacterial lineage from the lower ocean surface layer. Appl Environ Microbiol 63: 1441–1448.
Yoshinaga I, Fukami K, Ishida Y . (1991). Comparison of DNA and protein synthesis rates of bacterial assemblages between coral reef waters and pelagic waters in tropical ocean. Mar Ecol Prog Ser 76: 167–174.
Zhang R, Liu B, Lau SCK, Ki J-S, Qian P-Y . (2007). Particle-attached and free-living bacterial communities in a contrasting marine environment: Victoria Harbor, Hong Kong. FEMS Microbiol Ecol 61: 496–508.
Ziervogel K, Arnosti C . (2008). Polysaccharide hydrolysis in aggregates and free enzyme activity in aggregate-free seawater from the northeastern Gulf of Mexico. Environ Microbiol 10: 289–299.
Ziervogel K, Steen AD, Arnosti C . (2010). Changes in the spectrum and rates of extracellular enzyme activities in seawater following aggregate formation. Biogeosciences 7: 1007–1015.
Zimmerman AE, Martiny AC, Allison SD . (2013). Microdiversity of extracellular enzyme genes among sequenced prokaryotic genomes. ISME J 13: 1–13.
Zoppini A, Puddu A, Fazi S, Rosati M, Sist P . (2005). Extracellular enzyme activity and dynamics of bacterial community in mucilaginous aggregates of the northern Adriatic Sea. Sci Total Environ 353: 270–286.
Acknowledgements
Patrick Gibson and the captain and crew of the R/V Cape Hatteras kindly collected the offshore waters for us; Dr Steven Ross, chief scientist, gave permission to collect samples. We thank Sherif Ghobrial for assistance with sample processing in the lab. This project was supported by NSF (OCE-0848703 and -1332881 to CA). We are very grateful to Antje Boetius and two anonymous reviewers for their suggestions that considerably improved the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
D'Ambrosio, L., Ziervogel, K., MacGregor, B. et al. Composition and enzymatic function of particle-associated and free-living bacteria: a coastal/offshore comparison. ISME J 8, 2167–2179 (2014). https://doi.org/10.1038/ismej.2014.67
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2014.67
Keywords
This article is cited by
-
Anaerobic thiosulfate oxidation by the Roseobacter group is prevalent in marine biofilms
Nature Communications (2023)
-
Carbohydrates and carbohydrate degradation gene abundance and transcription in Atlantic waters of the Arctic
ISME Communications (2023)
-
Reduced bacterial mortality and enhanced viral productivity during sinking in the ocean
The ISME Journal (2022)
-
Ectohydrolytic enzyme activities of bacteria associated with Orbicella annularis coral
Coral Reefs (2021)
-
Marine Group-II archaea dominate particle-attached as well as free-living archaeal assemblages in the surface waters of Kongsfjorden, Svalbard, Arctic Ocean
Antonie van Leeuwenhoek (2021)