Abstract
Insects of the family Chironomidae, also known as chironomids, are distributed worldwide in a variety of water habitats. These insects display a wide range of tolerance toward metals and organic pollutions. Bacterial species known for their ability to degrade toxicants were identified from chironomid egg masses, leading to the hypothesis that bacteria may contribute to the survival of chironomids in polluted environments. To gain a better understanding of the bacterial communities that inhabit chironomids, the endogenous bacteria of egg masses and larvae were studied by 454-pyrosequencing. The microbial community of the egg masses was distinct from that of the larval stage, most likely due to the presence of one dominant bacterial Firmicutes taxon, which consisted of 28% of the total sequence reads from the larvae. This taxon may be an insect symbiont. The bacterial communities of both the egg masses and the larvae were found to include operational taxonomic units, which were closely related to species known as toxicant degraders. Furthermore, various bacterial species with the ability to detoxify metals were isolated from egg masses and larvae. Koch-like postulates were applied to demonstrate that chironomid endogenous bacterial species protect the insect from toxic heavy metals. We conclude that chironomids, which are considered pollution tolerant, are inhabited by stable endogenous bacterial communities that have a role in protecting their hosts from toxicants. This phenomenon, in which bacteria enable the continued existence of their host in hostile environments, may not be restricted only to chironomids.
Similar content being viewed by others
Introduction
Chironomids (Diptera: Chironomidae) inhabit virtually every type and condition of aquatic habitats (Armitage et al., 1995). They undergo a complete metamorphosis of three aquatic life stages (eggs, larvae and pupae), followed by the terrestrial adult stage, during which they do not feed or bite. Egg masses are laid on the surface of the water. Each gelatinous egg mass contains hundreds of eggs. Species of Chironomus display a wide range of tolerance toward metals and organic pollutions (Waterhouse and Farrel, 1985; Hellawell, 1986; Heliovaara and Vaisanen, 1993). Winner et al. (1980) found that chironomids made up ca. 80% of the total fauna in a stream heavily polluted by Cu, Cr and Zn, whereas they constituted less than 10% in an unpolluted section of the same stream. In another study, the response of benthic invertebrate communities to heavy metals in the Upper Arkansas River Basin, Colorado, revealed that sites located downstream of the primary sources of heavy metals were dominated by chironomids, whereas the clean water reference stations were dominated by mayflies (Ephemeroptera) (Clements, 1994). Despite the fact that chironomid tolerance toward pollution is well documented, their defensive mechanism in these contaminated environments is not fully understood (Armitage et al., 1995).
Chironomus sp. egg masses have recently been found to be natural reservoirs of Vibrio cholerae (Broza and Halpern, 2001; Halpern et al., 2003, 2004, 2006, 2007a; Broza et al., 2005; Halpern, 2010, 2011) and of pathogenic species of Aeromonas (Senderovich et al., 2008; Figueras et al., 2011; Beaz-Hidalgo et al., 2012). Although V. cholerae is a stable resident in chironomid egg masses, it makes up less than 0.5% of the endogenous bacterial community prevalent in this habitat (Halpern et al., 2007a).
When the bacterial community that inhabit the egg masses was studied using culturable methods, species from the following genera were cultured and identified: Acinetobacter, Aeromonas, Brachymonas, Exiguobacterium, Klebsiella, Leucobacter, Oceanobacillus, Paracoccus, Pseudomonas, Vibrio, Rheinheimera, Shewanella and Vibrio (Halpern et al., 2007a, 2007b, 2009a, 2009b; Raats and Halpern, 2007; Senderovich et al., 2008). Species of some of these genera are known for their ability to degrade toxicants, for example, Pseudomonas and Leucobacter (Halpern et al., 2007a, 2009a; Halpern, 2011). Similar results were obtained when denaturating gradient gel electrophoresis and cloning methods were used, indicating that the egg masses endogenous bacterial community is dominated by bacteria that are related to known detoxifying bacterial species (Senderovich and Halpern, 2012).
Following our findings (Halpern et al., 2007a; Senderovich and Halpern, 2012) regarding the toxicant degradation potential of the endogenous bacterial community in chironomid egg masses, we hypothesized that bacteria may contribute to the survival of chironomids larvae in polluted environments. To test this hypothesis, we explored for the first time, the endogenous bacterial communities that inhabit chironomid egg masses and larvae by using 454-pyrosequencing. In addition, by applying Koch postulates we were able to demonstrate that chironomid endogenous bacterial species indeed protect their host from toxicants.
Materials and methods
Sampling of chironomids for pyrosequencing analysis
Chironomid egg masses and larvae were sampled in northern Israel near Haifa at the Tivon waste stabilization pond (longitude 699, latitude 618), between September 2007 and October 2010. The egg masses were collected on styrofoam boards (25 × 25 cm) that were introduced into the water for 24 hours and served as artificial oviposition sites for adult females. Larvae were collected directly from the water. Immediately after collection, the egg masses and the larvae were brought into the laboratory. Each sample was washed five times and vortexed for 1 min in sterile saline water to remove any surface contaminants and any bacteria that were not attached to the egg mass or the larva. After washing, the egg masses and larvae were suspended in 2 ml 95% ethanol and kept at −20 °C until further examination.
DNA extraction and FLX-titanium pyrosequencing
Six egg masses and five larvae were crushed separately in 1 ml saline, using a sterile glass homogenizer. Total DNA was extracted using a DNA isolation kit (DNeasy Blood and Tissue, Qiagen, Hilden, Germany) according to the manufacturer's instructions and stored at −20 °C.
Microbial diversity was assessed in the six chironomid egg masses and five chironomid larvae by using the bacterial tag-encoded FLX-titanium amplicon pyrosequencing approach (Bailey et al., 2010; Pitta et al., 2010). This method is based upon similar principles as FLX-titanium amplicon pyrosequencing but utilizes titanium reagents and titanium procedures including a one-step PCR mixture of Hot Start and HotStar high fidelity taq polymerases, and amplicons originating and extending 350–450 bp from the 27 F region numbered in relation to Escherichia coli rRNA. The FLX-titanium amplicon pyrosequencing procedures were performed according to Wolcott et al., (2009) at the Research and Testing Laboratory (Lubbock, TX, USA) based upon RTL protocols (www.researchandtesting.com).
Sequence analysis
Amplicon lengths were 350–450 bp. A total of ca. 10 000 sequences per sample were obtained. Following sequencing, all failed sequence reads, low quality sequence ends, tags and chimeras were removed using the MOTHUR program (version 1.17.3) (Schloss et al., 2009). Taxonomic assignment of the sequences was achieved by the classify.seqs script using the trainset6_032010.rdp database file.
Sequences that overlapped over the longest span were aligned using the align.seqs script of MOTHUR with the SILVA compatible alignment database (Pruesse et al., 2007). The aligned data set was dereplicated to eliminate duplicate sequences. A distance matrix was calculated from the aligned sequences using the dist.seqs script, and operational taxonomic units (OTUs) defined by a 3% sequence distances level were assigned by hcluster script. We used an OTU rarefaction analysis to test whether the sampling regime adequately represented the bacterial diversity within each sample.
Principal coordinate analysis and analysis of molecular variance (AMOVA) were performed using this distance matrix. A newick-formatted tree that describes the dissimilarity (1-similarity) among multiple groups using the UPGMA (unweighted pair group method with arithmetic mean) algorithm and the distance between communities, was created by the tree.shared script.
Representative sequences of every OTU identified as Vibrio (accession numbers JQ431152–JQ431195) were separated from the other sequences by get.seqs script and were compared with those available in the GenBank database (http://www.ncbi.nlm.nih.gov), using the standard nucleotide–nucleotide BLAST program (BLASTN; http://www.ncbi.nlm.nih.gov), in order to identify these sequences to the species taxonomic level.
In order to find out more details about the members of the largest OTU that was present in every bacterial community associated with the larvae, representatives of this taxon were separated from other sequences by get.seqs script and taxonomically identified by classify.seqs script using the trainset6_032010.rdp database file, without applying a cutoff of bootstrap value in this command. This command enabled us to determine the closest relatives of the dominant OTU. Representative sequences of this OTU from each larval sample (accession numbers JQ431134–JQ431148) were compared with those available on the EzTaxon server (http://www.eztaxon.org/) (Chun et al., 2007) and PubMed database (http://www.ncbi.nlm.nih.gov/pubmed/) to determine their homology. A phylogenetic tree of the representative members of this large OTU from larval samples and their relative sequences from the databases was generated using the neighbor-joining method with NJPlot (MEGA 4.1; Tamura et al., 2007), based on alignments from CLUSTAL W.
Chironomid sampling for bacterial isolation and metal resistance bioassay
Egg masses and larvae were sampled and treated in order to remove all the bacteria that were not firmly attached as described above.
Isolation of metal-resistant bacteria from egg masses and larvae
Each egg mass and larva was crushed in 1 ml saline by using a sterile glass homogenizer. The homogenate was diluted and aliquots of 0.1 ml from each dilution were spread onto Luria agar (Luria broth, LB) (Himedia, Mumbai, India) and onto LB agar supplemented with 5 mM of one of the following compounds: potassium chromate (K2CrO4), lead(II) nitrate [Pb(NO3)2], copper(II) sulfate (CuSO4·5H2O) and zinc chloride (ZnCl2). Individual colonies were randomly picked and streaked on LB agar to obtain single colonies. Isolated colonies were subcultured at least four times before examination. The bacteria were identified by amplifying and sequencing a 1501-bp internal fragment of 16S rRNA gene in accordance to Senderovich et al., 2008. The obtained sequences were compared with those available in the EzTaxon server (http://www.eztaxon.org/) (Chun et al., 2007) to ascertain their closest relatives. The sequences were submitted to the GenBank database under the accession numbers: JQ582944–JQ582983. The resistance of the isolates to heavy metals were surveyed by cultivation on LB agar that was supplemented with increasing concentrations of one of the following compounds: K2CrO4, Pb(NO3)2, CuSO4, ZnCl2. The agar plates were incubated at 30 °C for 7 days.
Bacterial ability to detoxify heavy metals
Two bacterial endogenous species, Chromobacterium aquaticum strain YLNALPb2 (JQ582944) and Shewanella decolorationis strain YLZn3 (JQ582955), that were isolated from chironomid larvae, were tested for their ability to detoxify metals. The strains were cultured in LB supplemented with 0.5 mM Pb(NO3)2 or 5 mM K2CrO4, respectively. Each bacterial species was incubated at 30 °C for 3 days (with shaking). After incubation, cultures were centrifuged at 10 000 rcf for 15 min at 4 °C and supernatants were filtered through a 0.2 μm Millipore filter (Minisart hydrophilic syringe filter, Sartorius stedium biotech, Göttingen, Germany) and examined for the residual concentration of heavy metals. The concentration of lead was determined at the ‘Neve Ya’ar’ extension service laboratory (Ministry of Agriculture, Israel) in an Atomic Absorption apparatus (Varian industry, Spartanburg, SC, USA). Chromate-reducing activity was evaluated as the decrease in chromate concentration in the supernatant using the Cr(VI)-specific colorimetric reagent S-diphenylcarbazide (DPC), prepared in acetone/H2SO4 to minimize deterioration (Pattanapipitpaisal et al., 2001). Briefly, DPC (0.025 g) was dissolved in 9.67 ml acetone and 330 μl of 3 M H2SO4 was added. The reaction mixture was set up in an Eppendorf tube containing the following: 200 μl sample or standard potassium chromate solution, 400 μl 20 mM MOPS-NaOH buffer pH 7.0, 33 μl 3 M H2SO4, 40 μl 0.25% (w/v) DPC and 327 μl distilled water. Spectrophotometric measurements were carried out immediately at A540 (Pattanapipitpaisal et al., 2001).
Concentration of heavy metals in the sampling water
Lead and chromate concentrations were measured in the water from where the chironomids were sampled (Tivon waste stabilization pond), as described above.
Bioassay procedures
All the egg masses in the following experiments were freshly collected and treated by the washing procedure as described above. Egg masses were disinfected by immersing them for 3 min in 70% ethanol or in savior (cetrimide 0.5% w/v+ chlorexidine gluconate 0.05% w/v). The residue of the disinfectants was washed off in sterile distilled water. Untreated egg masses served as control. Each egg mass was transferred to a sterile tube containing 10 ml sterile distilled water for 48 h in order to let the larvae hatch. After hatching, 4–14 larvae were transferred to each well in a 24-well plates (the number of larvae per sample was not biased among treatments). Each well was supplemented with 2 ml of final solution as described below. In a preliminary experiment, Chromobacterium aquaticum (JQ582944) and Shewanella decolorationis (JQ582955) were isolated from larvae that hatched from untreated egg masses and survived in the presence of toxic metals (unpublished data).
Bioassay experiments with larvae after hatching
Experiment A (Pb)
Larvae that hatched from untreated and disinfected (savior) egg masses as described above, were exposed to 0.005 mM Pb(NO3)2. Each treatment was repeated six times. The experimental treatments were: (I) control larvae, exposed to 0.005 mM Pb(NO3)2; (II) savior disinfected larvae exposed to 0.005 mM Pb(NO3)2; (III) savior disinfected larvae exposed to 0.005 mM Pb(NO3)2 and supplemented with 106 cells per ml Chromobacterium aquaticum; (IV) control larvae in sterile water; and (V) disinfected larvae in sterile water (without metal supplementation).
Experiment B (Cr)
Larvae that hatched from untreated and disinfected (70% ethanol) egg masses as described above were exposed to 0.05 mM K2CrO4. Each treatment was repeated six times. The experimental treatments were: (I) control larvae, exposed to 0.05 mM K2CrO4; (II) ethanol disinfected larvae exposed to 0.05 mM K2CrO4; (III) ethanol disinfected larvae exposed to 0.05 mM K2CrO4 and supplemented with 106 cells per ml Shewanella decolorationis; (IV) control larvae in sterile water; and (V) disinfected larvae in sterile water (without metal supplementation).
In both experiments (A and B), the 24-well plates were incubated at room temperature for 48 h, after which the dead and the surviving larvae were counted. Larvae were considered dead if they did not respond to gentle prodding. Surviving larvae from treatment no. III were crushed in a sterile glass homogenizer and streaked on LB medium supplemented with 0.5 mM Pb(NO3)2 or K2CrO4 in order to check whether the added bacteria (Chromobacterium aquaticum or Shewanella decolorationis, respectively) can be reisolated as the major isolate. Suspected colonies were picked, subcultured four times on LB medium before examination and then were identified by 16S rRNA sequencing.
Statistical analysis of the bioassay results
The larval survival rates in experiment A (Pb) were negative for normal distribution (using SPSS, version 17.0) and thus, the differences between larval survival in the treatment groups were estimated using the Kruskal–Wallis nonparametric test. The larval survival rates in experiment B (Cr) were found to be normally distributed (using SPSS version 17.0); therefore, the differences between the larval survival in different treatment groups were estimated using the one-way-analysis of variance parametric test, followed by a post-hoc Tukey's test.
Chironomid identification
The taxonomic identification of chironomid species was performed by PCR amplification and sequencing of cytochrome oxidase subunit I gene (Folmer et al., 1994). The sequences were compared with those available in the GenBank databases (http://www.ncbi.nlm.nih.gov), using the standard nucleotide–nucleotide BLAST program (BLASTN; http://www.ncbi.nlm.nih.gov), to determine their homology. The sequences were submitted to the GenBank database under accession numbers JQ025709–JQ025718.
Results
Chironomid identification
All the egg masses and larvae that were sampled and analyzed in this study were identified as Chironomus transvaalensis.
454-pyrosequencing of 16S rRNA genes
The diversity of the bacterial communities in chironomid egg masses and larvae was surveyed by 454-pyrosequencing of the 16S rRNA genes. Initially, ca. 10 000 sequences per sample were obtained. However, after the low quality sequences were removed, 6600 sequences on average (±2500 s.d.) were analyzed per sample (72 900 sequences in total).
The richness of all the bacterial communities in the chironomid egg masses and larvae was estimated by rarefaction curves (Supplementary Figure S1). Bacterial community in the egg masses was more diverse (ca. 1000 OTUs per sample, analyzed at 3% dissimilarity level) than the community identified in the larvae (ca. 580 OTUs per sample). The average of Good's coverage for the OTU definition was 89% for the egg mass samples and 95% for the larva samples. The Chao richness estimator measured 1966 in the egg mass and 1079 in the larva samples. Each larva harbored a dominant OTU that made up about 28% of its microbial community on average. This dominant OTU may explain the differences in the richness of the bacterial communities in egg masses and larvae.
We compared the overall bacterial communities composition using the principal coordinate analysis. This analysis revealed a strong clustering of the samples according to the insect life cycle stage (egg mass and larva), indicating that egg masses and larvae have distinct microbial communities (Figure 1). When the distance between the bacterial communities associated with chironomid egg masses and larvae was examined by the UPGMA algorithm, it was found that the egg masses and the larvae clustered separately (Supplementary Figure S2). This observation was verified by analysis of molecular variance (AMOVA) which showed that significant differences were found between the microbial communities associated with the egg masses and the larvae (F1,9=4.3, P<0.001).
The diversity clustering of bacteria associated with six chironomid egg masses and five larvae (squares representing bacterial community associated with two larvae are one above the other). The first two principal coordinates (PC1 and PC2) from the principal coordinate analysis (PCoA) are plotted for each sample. The variance explained by the PCs is indicated in parentheses on the axes.
Taxonomic analysis
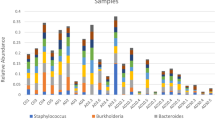
Taxonomic analysis of the sequences that were obtained by pyrosequencing revealed that the most prevalent phylum in the bacterial community associated with chironomid egg masses was Proteobacteria (ca. 58% of the sequences), whereas the most prevalent phylum in the larvae was Firmicutes (ca. 46% of the sequences) (Figure 2). Fifty-five percent of the sequences obtained from the egg masses and only 34% of the sequences from the larvae were classified to the genus taxonomic level. The dominant genera in the bacterial community associated with the egg masses were: Hydrogenophaga (8.1%), Pseudomonas (5.1%), Acinetobacter (4.9%), Arcobacter (4.2%), Acidovorax (3.8%), Aeromonas (1.6%), Aquabacterium (1.5%) and Dechloromonas (1.4%) (Table 1 and Supplementary Table S1). These genera were identified in each and every sampled egg mass (Table 1). The dominant genera in the bacterial community associated with the larvae were: Desulfovibrio (6.8%), Aeromonas (3.3%), Subdoligranulum (3.3%), Propionibacterium (1.9%), Aeromicrobium (1.8%), Thioflavicoccus (1.7%) and Hydrogenophaga (1.3%). These genera were present in all the larval samples (Table 1 and Supplementary Table S1).
Sequences that were identified as belonging to the Vibrio genus were found in all the egg mass samples and in one larva. Representative sequences of every Vibrio OTU were separated from other sequences and compared with those available in the GenBank databases for more accurate identification. These OTU representatives were identified as V. cholerae.
Forty percent of the bacterial community associated with the egg masses were found to belong to the same genera or species of various bacteria with known detoxification abilities, or to bacteria that were isolated from polluted sites. In the bacterial community associated with chironomid larvae, the bacteria that were closely related to detoxifying genera comprised 25% of the community (74% of sequences identified to the genus taxonomic level) (Table 1 and Supplementary Table S1).
The most dominant taxon in the bacterial community associated with the larvae was identified as unclassified Firmicutes, comprising 28% of the larval bacterial community on average. Representative sequences of this OTU from all larval samples were compared with those available in the databases to ascertain their closest relatives (Figure 3). These sequences were most closely related to Eubacterium sp. (AJ629069) and uncultured bacterium clones NkW01-038 (AB231068), PeM52 (AJ576412) and BOf1-07 (AB288878) (ca. 88% of sequence similarity). This taxon was not found in the bacterial community that was associated with the egg masses.
Representatives of the major OTU in the bacterial community associated with chironomid larvae, identified as unclassified Firmicutes (three sequences from each sample a–c were taken from five larva samples, L1–L5) (the accession numbers of these sequences are JQ431134–JQ431148). The phylogenetic tree represents the unclassified Firmicutes and their relatives from the EzTaxon server (http://www.eztaxon.org/) and PubMed database (http://www.ncbi.nlm.nih.gov/pubmed/). The sequence alignments were performed using the CLUSTAL W program, and the tree was generated using the neighbor-joining method with Kimura 2 parameter distances in MEGA 4.1 software (Tamura et al., 2007). Bootstrap values (from 1000 replicates) greater than 50% are shown at the branch points. The bar indicates a 2% sequence divergence.
Culturable metal-resistant isolates from egg masses and larvae
Bacterial strains resistant to toxic metals were isolated and identified from egg masses and larvae (Table 2, Supplementary Figure S3). The isolates were found to be resistant to more than one toxic metal. Some isolates exhibited a higher resistance to one certain metal in comparison with others (Table 2).
The ability of the chironomids' endogenous bacteria to detoxify heavy metals
Chromobacterium aquaticum strain YLNALPb2 and Shewanella decolorationis strain YLZn3, which have been isolated from larvae that hatched from untreated egg masses and survived in the presence of toxic metals (unpublished data), were tested for their ability to detoxify lead and hexavalent chromium, respectively. Both species showed the ability to reduce metal concentrations after an incubation period of three days. Ninety-four percent of Pb(NO3)2 was detoxified in the presence of Chromobacterium aquaticum strain YLNALPb2 (the Pb(NO3)2 concentration was reduced 17.3 times (STDEV 3.1)). Shewanella decolorationis strain YLZn3 neutralized 100% of the hexavalent chromium in the medium.
Heavy metal concentration at the sampling site of the chironomids
Lead and chromate were not detected in the water from which the chironomids were sampled.
Bioassay experiments
Two experiments were performed to assess the role of endogenous bacteria in protecting their chironomid host from toxicants. The experiments follow the rules of Koch-like postulates (a modified name for Koch's postulates is used here because the insects were disinfected before metal treatment) to demonstrate the role that endogenous bacteria have in their host.
Experiment A
Measuring the survival rate of disinfected larvae in the presence of lead and in the presence of lead supplemented with Chromobacterium aquaticum strain YLNALPb2.
Larvae that hatched from the egg masses without any disinfection treatment survived significantly better in lead-containing solution (77.6%) than the larvae that hatched from the egg masses that were disinfected with savior to remove their endogenous bacteria (30.9%). A supplementation of Chromobacterium aquaticum to the disinfected larvae significantly improved their survival rate (82.3%) (Figure 4a). The survival rates of larvae in the control treatments were higher than 93% (groups IV and V, larvae that hatched from nondisinfected and disinfected egg masses and were not exposed to lead nitrate). The differences between the survival rates of larvae that were exposed to different treatments were significant according to the Kruskal–Wallis nonparametric test (CHI2=13.674, df=2, P=0.001) (Figure 4a). Chromobacterium aquaticum was reisolated and identified from the treatment of disinfected larvae that survived in the presence of lead and the supplemented bacteria (according to Koch postulate no. 4).
Survival of disinfected larvae (mean±s.e.) in the presence of toxic lead (a) or hexavalent chromium (b) with and without the supplementation of chironomid endogenous bacteria. Untreated larvae which retained their endogenous bacteria survived significantly better in metal-contaminated environments compared with disinfected larvae. The addition of endogenous bacteria to the disinfected larvae (Chromobacterium aquaticum to the Pb(NO3)2 bioassay (a) and Shewanella decolorationis to the K2CrO4 bioassay (b)), improved their survival rate significantly. The added bacterial species were reisolated from the surviving larvae. The letters a or b in figure 4b represent the results of the post-hoc Tukey's test.
Experiment B
Measuring the survival rate of disinfected larvae in the presence of chromate and in the presence of chromate supplemented with Shewanella decolorationis strain YLZn3.
Larvae that hatched from the egg masses without any disinfection treatment and retained their endogenous bacteria, survived significantly better (85%) in the presence of chromate than the larvae that hatched from the egg masses that were disinfected with ethanol to remove their endogenous bacteria (45%). A supplement of Shewanella decolorationis to the disinfected larvae, significantly improved their survival rate in the presence of chromate (83%) (One-Way-analysis of variance (F4,25=9.97, P<0.05, R2=0.615) (Figure 4b)). The survival rates of larvae in the control treatment groups IV and V without toxic chromate, were high (97% and 75% respectively). The post-hoc Tukey's test demonstrated that the disinfected larvae that were exposed to chromate, survived in a significantly lower rate compared with all the other treatments (Figure 4b). Here too, as was demonstrated in experiment A, Shewanella decolorationis was isolated and identified from the surviving disinfected larvae that were supplemented with chromate and bacteria, demonstrating the protecting role of the bacteria in the larvae.
Discussion
Bacterial communities in chironomid egg masses and larvae
A stable endogenous bacterial community in chironomid egg masses and larvae was demonstrated by using the 454-pyrosequencing (Figure 1, Table 1 and Supplementary Table S1). Similar results for the association between the bacterial community and the egg masses were obtained by using denaturating gradient gel electrophoresis (Senderovich and Halpern, 2012). The PCR-denaturating gradient gel electrophoresis of egg masses that were sampled during a 5-month period revealed a unique banding pattern which didn't change significantly along the sampling period. To the best of our knowledge, this is the first study that explores bacterial community in chironomid larvae.
Bacterial community in egg masses was found to be more diverse and significantly different than that of the larvae (ca. 1000 OTUs in the egg mass samples, versus ca. 580 OTUs in larva samples) (Figures 1 and 2, Supplementary Figures S1 and S2). The differences between the two life stages may be due to a dominant unidentified Firmicutes OTU (28%) that was found in all the larval samples. The closest relatives of the dominant larval OTU (Figure 3) were: a clone (AB231068) identified from the gut wall of termites (Nakajima et al., 2006), a clone (AJ576412) identified from the hindgut of Pachnoda ephippiata larvae (Egert et al., 2003) and a clone (AB288878) identified from a fungus-growing termite (Shinzato et al., 2007). It has been suggested that these bacteria have symbiotic relationships with their hosts and may have a role in their nutrition digestion (Egert et al., 2003; Nakajima et al., 2006; Shinzato et al., 2007). More research is needed to verify the hypothesis regarding the symbiotic relationships between chironomids' larvae and this unclassified Firmicutes.
V. cholerae and Aeromonas as inhabitants of chironomids
V. cholerae has been found previously as a stable inhabitant of chironomid egg masses and flying adults (Broza and Halpern, 2001; Halpern et al., 2003, 2004, 2006, 2007a; Broza et al., 2005; Senderovich et al., 2008; Halpern, 2011). In the current study, V. cholerae was identified in all chironomid egg mass samples. It comprised a minor portion of the bacterial community in the egg mass (0.2% on average), as was found in a previous study (Halpern et al., 2007a). V. cholerae was identified only from one of the larval samples with relatively high abundance of 1.7% of the total bacterial community in this sample. A possible explanation for the absence of V. cholerae in the other four larval samples could be the presence of the dominant OTU in the larvae that masked the identification of species with relatively low abundances.
Chironomus sp. egg masses have also been implicated as a natural reservoir of pathogenic Aeromonas species (Senderovich et al., 2008; Figueras et al., 2011; Beaz-Hidalgo et al., 2012). In the current study, Aeromonas was identified in all the egg masses and the larval samples, comprising an average of 1.6% and 3.3% of the bacterial communities in the egg masses and the larvae, respectively (Table 1). These results again demonstrate that Aeromonas is a stable resident in the chironomid niche. Some of the Aeromonas isolates have previously been found to degrade chironomid egg masses into individual eggs, which might affect egg's hatching (Senderovich et al., 2008). In the current study, culturable Aeromonas isolates showed the ability to detoxify toxic metals (Table 2). These properties of Aeromonas isolates should be further investigated to understand the relationships between Aeromonas and chironomids.
The possible protective role of endogenous bacteria in chironomids
About 40% of the bacterial community associated with the egg masses and 25% of the bacterial community associated with the larvae were found to be closely related to bacteria that are known as degraders of various toxicants (Table 1 and Supplementary Table S1). The total percentage of detoxifying bacteria in the bacterial community associated with the larvae is smaller than in those associated with the egg masses. This is most likely because of the overabundance of the dominant larval OTU (Figure 3). Nevertheless, the most prevalent detoxifying genera that were found in all the egg mass samples (Hydrogenophaga, Acidovorax, Dechloromonas, Comamonas, Diaphorobacter, Pseudomonas, Acinetobacter and Aeromonas) were also present in almost all the larval samples (Table 1). OTUs belonging to the genus Leucobacter were found in all the egg mass samples and in three out of five larval samples (Supplementary Table S1). Recently, we have isolated a novel species of Leucobacter from a chironomid egg mass, which was identified and characterized by us as Leucobacter chironomi sp. nov. (Halpern et al., 2009a). This species displayed high chromate resistance (18.0 mM Cr(VI)) although it was isolated on LB agar plate and from an environment that was not polluted with toxic chromate (Halpern et al., 2009a).
Chironomids are considered pollution tolerant (Winner et al., 1980; Cranston, 1995; Thorne and Williams, 1997; Richardson and Kiffney, 2000). It has been shown that chironomids are able to acclimate and adapt to metals in their environment (Miller and Hendricks, 1996; Groenendijk et al., 2002). However, the mechanisms for these adaptations are not clear. It was suggested that they excrete the metals (Postma et al., 1996; Groenendijk et al., 1999) or bind them with metal-binding proteins (Yamamura et al., 1983). Here we demonstrate that endogenous bacteria have a role in protecting the insect from pollutants. Culturable endogenous bacteria that showed resistance to toxic metals were isolated from chironomid egg masses and larvae (Table 2, Supplementary Figure S3). Moreover, Koch-like postulates were applied to demonstrate that specific bacterial species that were isolated from the larvae have a role in protecting chironomid larvae from toxic metals.
Application of Koch-like postulates to provide evidence for the role of endogenous bacteria in chironomids
Chromium is one of the major sources of environmental pollution (Komori et al., 1990; Desh and Gupta, 1991). It can permeate through biological membranes and interact with intracellular proteins and nucleic acids (Horitsu et al., 1978). Lead is a hazardous metal of environmental concern with high persistency. This metal has no biological function and can induce, directly or indirectly, various morphological, physiological and biochemical dysfunctions in different organisms (Shahid et al., 2012).
Chromobacterium aquaticum strain YLNALPb2 and Shewanella decolorationis strain YLZn3 that were isolated from chironomid larvae, reduced toxic lead (up to 94%) and hexavalent chromium (up to 100%) concentrations, respectively. The role of these species in chironomids was demonstrated in bioassays that tested the survival rate of chironomid larvae in environments containing heavy metals. Untreated larvae that contained endogenous bacteria, survived significantly better in lead nitrate and chromate containing environments, compared with disinfected larvae (Figures 4a and b). The supplementation of Chromobacterium aquaticum or Shewanella decolorationis to the disinfected larvae in lead or chromate containing environments, respectively, improved the survival rate of the disinfected larvae, significantly (Figures 4a and b). Following Koch's postulates, Chromobacterium aquaticum and Shewanella decolorationis that were added to the disinfected larvae were reisolated from the larvae that survived in the bioassays (Figures 4a and b).
The role of endogenous bacterial community in a host
Mutualism (in which both organisms benefit) and commensalism (where one organism benefits without affecting the other), between prokaryotes and eukaryotes are well-known phenomena (for example, Friedrich et al., 2001; Rohwer et al., 2002; Fraune and Bosch, 2007). These associations between prokaryotes and their larger hosts were recently demonstrated in the hologenome theory of evolution. According to this theory, the holobiont (the host together with its endogenous microorganisms) is a unit of selection in evolution (Rosenberg et al., 2007; Zilber-Rosenberg and Rosenberg, 2008; Rosenberg and Zilber-Rosenberg, 2011). Here we demonstrate an example for this theory. Chironomids, known as pollution inhabitants, are protected from toxic metals by endogenous bacteria, which help them to survive in metal-contaminated environments. This association is most likely a result of coevolution between the host and its endogenous microorganisms (holobiont). This relationship is of special interest as it could indicate that revision of traditional views of metal accumulation through animal trophic levels, may be too simplistic. Many studies have been conducted in order to better understand the accumulation patterns of metals in different species of invertebrates and fish and between various aquatic systems (freshwater and marine) (reviewed in Wang and Rainbow, 2008). None of these studies considered the potential detoxification activity of bacteria in this process. Recently, Selvin et al. (2009) found that a marine sponge Fasciospongia cavernosa, is inhabited by bacterial species that showed resistance against Cd and Hg. In another study, multimetal resistance was observed in a bacterial population isolated from coral tissues in coastal water, Indonesia (Sabdono et al., 2012). These examples demonstrate that, indeed, metal-resistant bacteria inhabit aquatic invertebrates and thus, may help them to survive in metal-polluted environments as was demonstrated here for chironomids.
Conclusions
Using Koch-like postulates, we demonstrated that endogenous bacteria in chironomids have a role in protecting their host from toxic heavy metals. The 454-pyrosequencing of the bacterial communities in chironomids implies that other endogenous species have the potential of protecting the insect from a huge verity of toxicants (for example, other heavy metals, pesticides and aromatic hydrocarbons) (Table 1 and Supplementary Table S1). This phenomenon, in which bacteria enable the continued existence of their host in hostile environments, may not be restricted only to chironomids.
Accession codes
Accessions
GenBank/EMBL/DDBJ
References
Armitage P, Cranston PS, Pinder LCV . (1995) The Chironomidae: the Biology and Ecology of Non-biting Midges. Chapman and Hall: London, UK.
Bailey MT, Walton JC, Dowd SE, Weil ZM, Nelson RJ . (2010). Photoperiod modulates gut bacteria composition in male siberian hamsters (Phodopus sungorus). Brain, Behav Immun 24: 577–584.
Beaz-Hidalgo R, Shakèd T, Laviad S, Halpern M, Figueras M . (2012). Chironomid egg masses harbour the clinical species Aeromonas taiwanensis and Aeromonas sanarellii. FEMS Microbiol Lett 337: 48–54.
Broza M, Halpern M . (2001). Chironomid egg masses and Vibrio cholerae. Nature 412: 40.
Broza M, Gancz H, Halpern M, Kashi Y . (2005). Adult non-biting midges: possible windborne carriers of Vibrio cholerae. Environ Microbiol l7: 576–585.
Chakraborty R, O’Connor SM, Chan E, Coates JD . (2005). Anaerobic degradation of benzene, toluene, ethylbenzene, and xylene ompounds by Dechloromonas strain RCB. Appl Environ Microbiol 71: 8649–8655.
Chaillan F, Le Flèche A, Bury E, Phantavong YH, Grimont P, Saliot A et al (2004). Identification and biodegradation potential of tropical aerobic hydrocarbon-degrading microorganisms. Res Microbiol 155: 584–595.
Chang Y-C, Ikeutsu K, Toyama T, Choi DB, Kikuchi S . (2011). Isolation and characterization of tetrachloroethylene- and cis-1,2-dichloroethylene-dechlorinating propionibacteria. J Ind Microbiol Biotechnol 38: 1667–1677.
Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK et al (2007). EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J System Evol Microbiol 57: 2259–2261.
Clements WH . (1994). Benthic invertebrate community responses to heavy metals in the Upper Arkansas River Basin, Colorado. J North Am Benlhol Soc 13: 30–44.
Cranston PS . (1995). Introduction. In: The Chironomidae: The Biology and Ecology of Non-biting Midges Armitage PD, Cranston PS, Pinder LCV, (eds). Chapman and Hall: London, UK, pp 1–7.
Cruz A, Caetano T, Suzuki S, Mendo S . (2007). Aeromonas veronii, a tributyltin (TBT)-degrading bacterium isolated from an estuarine environment, Ria de Aveiro in Portugal. Mar Environ Res 64: 639–650.
Desh D, Gupta AK . (1991). Hexavalent chromium removal from waste water. Indian J Environ Health 33: 297–305.
Egert M, Wagnerl B, Lemke T, Brune A, Friedrich M . (2003). Microbial community structure in midgut and hindgut of the humus-feeding larva of Pachnoda ephippiata (Coleoptera: Scarabaeidae). Appl Environ Microbiol 69: 6659–6668.
Figueras MJ, Beaz-Hidalgo R, Senderovich Y, Laviad S, Halpern M . (2011). Re-identification of Aeromonas isolates from chironomid egg masses as the potential pathogenic bacteria Aeromonas aquariorum. Environ Microbiol Rep 3: 239–244.
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R . (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3: 294–299.
Fraune S, Bosch TCG . (2007). Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc Natl Acad Sci USA 104: 13146–13151.
Friedrich AB, Fischer I, Proksch P, Hacker J, Hentschel U . (2001). Temporal variation of the microbial community associated with the mediterranean sponge Aplysina aerophoba. FEMS Microbiol Ecol 38: 105–113.
Groenendijk D, Lücker SMG, Plans M, Kraak MHS, Admiraal W . (2002). Dynamics of metal adaptation in riverine chironomids. Environ Pollut 117: 101–109.
Groenendijk D, van Kraak MHS, Admiraal W . (1999). Efficient shedding of accumulated metals during metamorphosis in metal adapted populations of the midge Chironomus riparius. Environ Toxicol Chem 18: 1225–1231.
Halpern M . (2010). Novel insights into hemagglutinin protease (HAP) gene regulation in Vibrio cholerae. Mol Ecol 19: 4108–4112.
Halpern M . (2011). Chironomids and Vibrio cholerae. In: Beneficial Microorganisms in Multicultural Life Forms Rosenberg E, Gophna U, (eds). Springer-Verlag: Berlin Heidelberg, Germany, pp 43–56.
Halpern M, Gancz H, Broza M, Kashi Y . (2003). Vibrio cholerae hemagglutinin/protease degrades chironomid egg masses. Appl Environ Microbiol 69: 4200–4204.
Halpern M, Broza YB, Mittler S, Arakawa E, Broza M . (2004). Chironomid egg masses as a natural reservoir of Vibrio cholerae non-O1 and non-O139 in freshwater habitats. Microb Ecol 47: 341–349.
Halpern M, Raats D, Lavion R, Mittler S . (2006). Dependent population dynamics between chironomids (non-biting midges) and Vibrio cholerae. FEMS Microbiol Ecol 55: 98–104.
Halpern M, Landsberg O, Raats D, Rosenberg E . (2007a). Culturable and VBNC Vibrio cholerae: interactions with chironomid egg masses and their bacterial population. Microb Ecol 53: 285–293.
Halpern M, Senderovich Y, Snir S . (2007b). Rheinheimera chironomi sp. nov., isolated from a chironomid (Diptera; Chironomidae) egg mass. Int J Syst Evol Microbiol 57: 1872–1875.
Halpern M, Shaked T, Pukall R, Schumann P . (2009a). Leucobacter chironomi sp. nov., a chromate resistant bacterium isolated from a chironomid egg mass. Int J Syst Evol Microbiol 59: 665–670.
Halpern M, Shaked T, Schumann P . (2009b). Brachymonas chironomi sp. nov., isolated from a chironomid egg mass, and emended description of the genus Brachymonas. Int J Syst Evol Microbiol 59: 3025–3029.
Heliovaara K, Vaisanen R . (1993) Insects and Pollution. CRC Press: Boca Raton, FL, USA.
Hellawell JM . (1986) Biological Indicators of freshwater Pollution and Environmental Management. Elsevier Applied Science Publishers: London, UK, pp 546.
Horitsu H, Nishida H, Kato H, Tomoyeda M . (1978). Isolation of potassium chromate tolerant bacterium and chromate uptake by the bacterium. Agric Biol Chem 42: 2037–2043.
Klankeo P, Nopcharoenkul W, Pinyakong O . (2009). Two novel pyrene-degrading Diaphorobacter sp. and Pseudoxanthomonas sp. isolated from soil. J Biosci Bioeng 108: 488–495.
Komori K, Wang P, Toda K, Ohtake H . (1990). A method for removal of toxic chromium using dialysis-sac cultures of’ a chromate reducing strain of Enterobacter cloacae. Appl Microbiol Biotechnol 33: 117–119.
Lambo AJ, Patel TR . (2007). Biodegradation of polychlorinated biphenyls in Aroclor 1232 and production of metabolites from 2,4,4′-trichlorobiphenyl at low temperature by psychrotolerant Hydrogenophaga sp. strain IA3-A. J Appl Microbiol 102: 1318–1329.
Lee CM . (1994). Effects of immobilized cells on the biodegradation of chlorinated phenols. Water Sci Technol 30: 87–90.
Michel C, Brugna M, Aubert C, Bernadac A, Bruschi M . (2001). Enzymatic reduction of chromate: comparative studies using sulfate-reducing bacteria. Key role of polyheme cytochromes c and hydrogenases. Appl Microbiol Biotechnol 55: 95–100.
Miller MP, Hendricks AC . (1996). Zinc resistance in Chironomus riparius: evidence for physiological and genetic components. J North Amer Benthol Soc 15: 106–116.
Nakajima H, Hongoh Y, Noda S, Yoshida Y, Yusami R, Kudo T et al (2006). Phylogenetic and morphological diversity of bacteroidales members associated with the gut wall of termites. Biosci Biotechnol Biochem 1: 211–218.
Ohtsubo Y, Goto H, Nagata Y, Kudo T, Tsuda M . (2006). Identification of a response regulator gene for catabolite control from a PCB-degrading b-proteobacteria, Acidovorax sp. KKS102. Mol Microbiol 60: 1563–1575.
Okeke BC . (2008). Bioremoval of hexavalent chromium from water by a salt tolerant bacterium, Exiguobacterium sp. GS1. J Ind Microbiol Biotechnol 35: 1571–1579.
Pattanapipitpaisal P, Brown NL, Macaskie LE . (2001). Chromate reduction and 16S rRNA identification of bacteria isolated from a Cr(VI)-contaminated site. Appl Microbiol Biotechnol 57: 257–261.
Pitta DW, Pinchak E, Dowd SE, Osterstock J, Gontcharova V, Youn E et al (2010). Rumen bacterial diversity dynamics associated with changing from Bermuda grass hay to grazed winter wheat diets. Microb Ecol 59: 511–522.
Postma JF, Van Nugteren P, Buckert-De Jong MB . (1996). Increased cadmium excretion in metal-adapted populations of the midge Chironomus riparius (Diptera). Environ Toxicol Chem 15: 332–339.
Pruesse E, Quast C, Knittel K, Fuchs B, Ludwig W, Peplies J et al (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196.
Raats D, Halpern M . (2007). Oceanobacillus chironomi sp. nov., a halotolerant and facultative alkaliphilic species isolated from a chironomid egg mass. Int J Syst Evol Microbiol 57: 255–259.
Richardson JS, Kiffney PM . (2000). Responses of a macroinvertebrate community from a pristine, southern British Columbia, Canada, stream to metals in experimental mesocosms. Environ Toxicol Chem 19: 736–743.
Rohwer F, Seguritan V, Azam F, Knowlton N . (2002). Diversity and distribution of coral-associated bacteria. Mar Ecol: Prog Ser 243: 1–10.
Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I . (2007). The role of microorganisms in coral health, disease and evolution. Nature Rev Microbiol 5: 355–362.
Rosenberg E, Zilber-Rosenberg I . (2011). Symbiosis and development: the hologenome concept. Birth Defects Res C Embryo Today 93: 56–66.
Sabdono A, Radjasa OK, Utomo HS . (2012). Screening of multi-metal resistances in a bacterial population isolated from coral tissues of central Java coastal waters, Indonesia. Int J Oceanog Marine Ecol Syst 1: 11–23.
Samanta S, Singh OV, Jain RK . (2002). Polycyclic aromatic hydrocarbons: environmental pollution and bioremediation. Trends Biotechnol 20: 243–248.
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollist EB et al (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541.
Selvin J, Shanmugha Priya S, Seghal Kiran G, Thangavelu T, Sapna Bai N . (2009). Sponge-associated marine bacteria as indicators of heavy metal pollution. Microbiol Res 164: 352–363.
Senderovich Y, Gershtein Y, Halewa E, Halpern M . (2008). Vibrio cholerae and Aeromonas; do they share a mutual host? ISME J 2: 276–283.
Senderovich Y, Halpern M . (2012). Bacterial community composition associated with chironomid egg masses. J Insect Sci 12: 149.
Shahid M, Pinelli E, Dumat C . (2012). Review of Pb availability and toxicity to plants in relation with metal speciation; role of synthetic and natural organic ligands. J Hazard Mat 15: 219–220.
Shinzato N, Muramatsu M, Matsui T, Watanabe Y . (2007). Phylogenetic analysis of the gut bacterial microflora of the fungus-growing termite Odontotermes formosanus. Biosci Biotechnol Biochem 4: 906–915.
Sylvestre M . (1995). Biphenyl/chlorobiphenyls catabolic pathway of Comamonas testosteroni B-356: prospect for use in bioremediation. Int Biodeterior Biodegrad 35: 189–211.
Tamura K, Dudley J, Nei M, Kumar S . (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599.
Thorne RSJ, Williams WP . (1997). The response of benthic macroinvertebrates to pollution in developing countries: a multimetric system of bioassessment. Freshwater Biol 37: 671–686.
Wang WX, Rainbow PS . (2008). Comparative approaches to understand metal bioaccumulation in aquatic animals. Comp Biochem Physiol C Toxicol Pharmacol 148: 315–323.
Waterhouse IC, Farrell MP . (1985). Identifying pollution related changes in chironomid communities as a function of taxonomic rank. Can J Fish Aquat Sci 42: 406–413.
Williams PA, Sayers JR . (1994). The evolution of pathways for aromatic hydrocarbon oxidation in Pseudomonas. Biodegradation 5: 195–217.
Winner RW, Bossel MW, Farrel MP . (1980). Insect community structure as an index of heavy-metal pollution in lotic ecosystems. Can J Fish Aquat Sci 37: 647–655.
Wolcott RD, Gontcharova V, Sun Y, Dowd SE . (2009). Evaluation of the bacterial diversity among and within individual venous leg ulcers using bacterial tag-encoded FLX and titanium amplicon pyrosequencing and metagenomic approaches. BMC Microbiol 9: 226.
Yamamura M, Suzuki KT, Hatakeyama S, Kubota K . (1983). Tolerance to cadmium and cadmium-binding proteins induced in the midge larva, Chironomus yoshimatsui (Diptera, Chironomidae). Comp Biochem Physiol C Chomp Pharmacol 75: 21–24.
Zilber-Rosenberg I, Rosenberg E . (2008). Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol Rev 32: 723–735.
Acknowledgements
This study was partially supported by a grant from the US Civilian Research and Development Foundation (CRDF grant no. ILB1-7045-HA). We thank Sivan Laviad and Tamar Shakéd for their technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Senderovich, Y., Halpern, M. The protective role of endogenous bacterial communities in chironomid egg masses and larvae. ISME J 7, 2147–2158 (2013). https://doi.org/10.1038/ismej.2013.100
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2013.100
Keywords
This article is cited by
-
Honey as a sustainable indicator of heavy metals in tropical rainforest vegetation zone: an early warning monitoring approach
International Journal of Tropical Insect Science (2023)
-
Characterization of tasar silkworm Antheraea mylitta drury (Saturniidae: Lepidoptera) midgut bacterial symbionts through metagenomic analysis
International Journal of Tropical Insect Science (2023)
-
Chironomus riparius Larval Gut Bacteriobiota and Its Potential in Microplastic Degradation
Microbial Ecology (2023)
-
Species Identity, Life History, and Geographic Distance Influence Gut Bacterial Communities in Lab-Reared and European Field-Collected Culicoides Biting midges
Microbial Ecology (2022)
-
Stress response of Lymantria dispar asiatica (Lepidoptera: Erebidae) larvae and its gut microbiota to manganese ion
Journal of Forestry Research (2021)