Abstract
Despite its immense size, logistical and methodological constraints have largely limited microbiological investigations of the subseafloor basement biosphere. In this study, a unique sampling system was used to collect fluids from the subseafloor basaltic crust via a Circulation Obviation Retrofit Kit (CORK) observatory at Integrated Ocean Drilling Program borehole 1301A, located at a depth of 2667 m in the Pacific Ocean on the eastern flank of the Juan de Fuca Ridge. Here, a fluid delivery line directly accesses a 3.5 million years old basalt-hosted basement aquifer, overlaid by 262 m of sediment, which serves as a barrier to direct exchange with bottom seawater. At an average of 1.2 × 104 cells ml−1, microorganisms in borehole fluids were nearly an order of magnitude less abundant than in surrounding bottom seawater. Ribosomal RNA genes were characterized from basement fluids, providing the first snapshots of microbial community structure using a high-integrity fluid delivery line. Interestingly, microbial communities retrieved from different CORKs (1026B and 1301A) nearly a decade apart shared major community members, consistent with hydrogeological connectivity. However, over three sampling years, the dominant gene clone lineage changed from relatives of Candidatus Desulforudis audaxviator within the bacterial phylum Firmicutes in 2008 to the Miscellaneous Crenarchaeotic Group in 2009 and a lineage within the JTB35 group of Gammaproteobacteria in 2010, and statistically significant variation in microbial community structure was observed. The enumeration of different phylogenetic groups of cells within borehole 1301A fluids supported our observation that the deep subsurface microbial community was temporally dynamic.
Similar content being viewed by others
Introduction
It is now generally accepted that a subseafloor biosphere extends throughout the enormous volume of sediments and basement basalt that make up the global system of mid-ocean ridge spreading centers, flanks and ocean basins (for example, Gold, 1992; Parkes et al., 1994; Fisk et al., 1998; Bach and Edwards, 2003; Cowen et al., 2003; Schrenk et al., 2010; Orcutt et al., 2011a). Of the different oceanic subsurface environments, the porous, uppermost igneous crust originating from the mid-ocean ridge spreading centers is likely to be the most suitable for microbial life, because it is the locus of extensive hydrothermal circulation (Baross et al., 2004). Vigorous seawater entrainment into young, <10 million years old (Ma) ridge flanks actively injects a chemical disequilibrium (generally in the form of terminal electron acceptors) into the deep ocean crust, fueling redox-active processes involving iron and sulfur cycling. This disequilibrium is thought to sustain a substantial subseafloor microbial ecosystem (Bach and Edwards, 2003).
Despite the presumed pervasiveness of the basalt-hosted deep subsurface biosphere, a thick layer of sediment that significantly restricts sampling opportunities covers much of the oceanic basement of the mid-ocean ridge flanks and ocean basins. However, seafloor instrumentation platforms known as Circulation Obviation Retrofit Kit (CORK) observatories (Davis et al., 1992; Edwards et al., 2011) affixed to the Ocean Drilling Program (ODP) and Integrated Ocean Drilling Program (IODP) boreholes (Davis and Becker, 2001; Fisher et al., 2005a) provide a rare opportunity to conduct in situ geophysical studies and sample fluids from the sediment-covered basement rock. By channeling fluids through dedicated microbiological and geochemical sampling lines, natural crustal fluids originating from the deep subsurface can be retrieved at the seafloor for subsequent interrogation.
Several boreholes drilled in the seafloor on the eastern flank of the Juan de Fuca Ridge (JdFR) in the northeastern Pacific Ocean are equipped with CORK observatories, providing access to 1.2 to 3.5 Ma crust that is covered by up to several hundred meters (m) of sediment. This sediment results in an impermeable seal between bottom seawater and the basement rock (Embley et al., 1983). In this particular region of seafloor, nearby rocky seamounts protruding through the sediments combine with high basement permeability to provide hydrogeological exchange between hydrothermal recharge and discharge points over distances greater than 50 km (Fisher et al., 2003). These geological features allow deep seawater to be entrained into the ocean basement and to evolve during its passage through the relatively warm and chemically reducing basement rock environment (Cowen, 2004). The biological and chemical characteristics of crustal fluids collected via CORK observatories tapping into basement fluid recharge zones likely reflect the response of the in situ basement microbial community to an influx of terminal electron acceptors (Wheat et al., 2010; Lin et al., 2012).
In 2003, Cowen et al. used an early-generation CORK observatory to collect basement fluids originating deep within the JdFR borehole 1026B, to conduct one of the first microbiological studies of young ridge flank crustal fluids. A passive flow ‘BioColumn’ device was connected in series to the CORK spigot to channel discharging fluids through a filter membrane. Subsequently, Huber et al. (2006) sampled fluids emanating from borehole 1026B, as well as those exiting Baby Bare Seamount (an exposed basaltic seamount nearby), using an in situ filtration system to collect microbial biomass. Subsurface fluids exiting the seamount were sampled by Huber et al. (2006) using stainless steel probes inserted through thin sediment and into the basement rock. Although young ridge flank fluid microbial diversity has been assessed using CORK-enabled in situ enrichment techniques (Smith et al., 2011; Orcutt et al., 2011b) and biofilm sampling of a retrieved CORK exposed to ridge flank crustal fluids (Nakagawa et al., 2006), the two samples collected from borehole 1026B and samples collected from Baby Bare seamount have provided the lone assessments of planktonic microbial community structure within young ridge flank fluids.
Recent technological advances in CORK-assisted basement fluid sampling permit unprecedented access to basement fluids. Relatively inert stainless steel fluid delivery lines running exterior to the CORK casing are featured on a new generation of CORK observatories deployed on both pre-existing (1026B) and new (1301A) boreholes (Fisher et al., 2005b) in an effort to sample deeply buried basement fluids while controlling contamination and minimizing biofouling effects noted in previous studies (Cowen et al., 2003). In this study, a pump with titanium- and Teflon-wetted parts was used to draw basement fluids up the fluid delivery line and through the CORK spigot, permitting thorough flushing of the fluid delivery lines before the collection of basement fluids (Cowen et al., 2012; Lin et al., 2012). The upgraded materials and extensive flushing of the fluid delivery line facilitate high-integrity sampling of the crustal fluids, providing for an accurate assessment of in situ microbial community characteristics from the sediment-covered basement rock of the ridge flanks. Here, we describe the use of these improvements to retrieve crustal fluids from CORK 1301A in the summers of 2008–2010. Small subunit ribosomal RNA (SSU rRNA) gene cloning and sequencing and epifluorescence microscopy were used to provide insight into the diversity of microbial communities inhabiting the igneous crustal biosphere.
Materials and methods
Sample collection
During R/V Atlantis cruises ATL15_35 (28 July 2008–13 August 2008), ATL15_51 (20 August 2009–6 September 2009) and ATL15_66 (13 June 2010–1 July 2010), basement crustal fluids were collected from a CORK-II borehole observatory at IODP borehole 1301A located 101 km east of the JdFR spreading center (47°45.209′N, 127°45.833′W; Figure 1). Fluids were sampled from the microbiological and geochemical sampling lines associated with the CORK-II installation (Fisher et al., 2005a). Borehole 1301A penetrates 262 m of sediments and another 108 m into ∼3.5 Ma basement rock (Table 1). A deep-sea pumping system was used that was improved significantly over the course of the study, becoming more powerful and incorporating a non-contaminating titanium and Teflon pump head and complementary in-line sensors (Lin et al., 2012; Figure 1c). Fluid samples were ultimately collected in custom-made 60-l acid-washed Tedlar bags (MiDan Co., Chino, CA, USA) protected by a high-density polyethylene box (Figure 1). To minimize the introduction of chemical and microbial contaminants during sampling, acid-washed tubing and sampling bags were employed. In 2008, sample bags were filled after flushing the fluid delivery line with three times their volume of borehole fluids, and elevated fluid temperatures were observed. In 2009 and 2010, fluids were collected into sample bags after the fluid temperature had risen and stabilized at ∼20 °C for 1 h. Subsurface samples described herein were collected during human-occupied vehicle Alvin dives 4434 (2008) and 4532 (2009), and remote-operated vehicle Jason II dive 497 (2010).
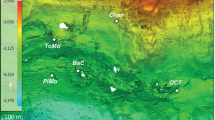
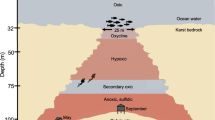
(a) Location of CORK observatory sampling sites on the Juan de Fuca Ridge flank, Pacific Ocean. (b) Schematic diagram of CORK located at borehole 1301A. Borehole crustal fluids were sampled from the exit valve of the fluid delivery line. (c) Photos of the fluid sampling device used in 2008, when a SeaBird pump (SBE) was used to divert fluids into the large volume bag sampler (LVBS), and 2009 and 2010, when a mobile pump sampler (MPS) was used to draw fluids and delivery into the LVBS. Figure modified from Lin et al. (2012).
See Supplementary Information for details regarding sample preparation, DNA extraction, cloning and sequencing of SSU rRNA genes, sequence analysis, oligonucleotide probe design and microscopic analysis.
Results
Physical and chemical characteristics of borehole 1301A fluid
The chemical characteristics of crustal fluids from borehole 1301A retrieved concomitantly with the samples analyzed in this study have been described previously (Lin et al., 2012). High-integrity fluid samples were of near neutral pH (7.4–7.5), depleted in dissolved organic carbon, carbon dioxide, nitrate, sulfate and phosphate, and enriched in ammonium, silicate, calcium, and ferric and ferrous iron (Table 2). Magnesium concentrations serve as an indicator of hydrothermally altered subsurface fluids (Mottl and Wheat, 1994); the high-integrity borehole fluid samples analyzed in this study were selected on the basis of depleted magnesium concentrations relative to bottom seawater (Table 2).
Microbial community structure
Universal oligonucleotide PCR primers were used to clone and sequence SSU rRNA genes from one high-integrity crustal fluid sample collected from borehole 1301A in each of three consecutive field seasons (2008–2010). A total of 238 (2008), 353 (2009) and 191 (2010) environmental gene clones were sequenced (Supplementary Table S1). In addition, 72 (2008), 363 (2009) and 176 (2010) gene clones were sequenced from libraries generated from bottom seawater collected in the vicinity of borehole 1301A via conductivity, temperature and depth (CTD) rosette, and 95 environmental gene clones were sequenced from a sample of bottom seawater collected in the vicinity of borehole 1301A using a Niskin bottle attached to the remote-operated vehicle Jason II in 2010 (Supplementary Table S2). Borehole fluid sample microbial communities were analyzed using a variety of α-diversity calculators and operational taxonomic units defined at 99% and 97% SSU rRNA gene sequence similarity (Supplementary Table S3). The Shannon diversity index showed an increasing trend over the 3-year sampling period (Supplementary Table S3), indicating an increase in community diversity over time. Rarefaction curves generated using the same operational taxonomic unit definitions were for the most part steeply sloping, indicating that the clone libraries are undersampled (Supplementary Figure S1).
In each of the 3 years, the microbial community structure within borehole 1301A fluid was significantly different than that of its corresponding bottom seawater control(s), based on the UniFrac weighted and unweighted tests (Supplementary Table S4). In general, borehole fluid microbial communities were dominated by clones related to microorganisms harboring physiological attributes consistent with the physical and chemical conditions of this environment (for example, thermophiles, anaerobes, sulfate-reducers, and so on), or related to SSU rRNA gene sequences previously recovered from related environments (Supplementary Table S1). In contrast, seawater samples were generally dominated by environmental clones related to bacterial and archaeal lineages previously recovered from seawater (Supplementary Table S2). On the basis of the operational taxonomic unit clustering, bottom seawater and borehole fluid microbial communities shared a minor component of their respective diversity (Supplementary Figure S2). The three borehole fluid clone libraries were also significantly different between years (Supplementary Table S4), and were each dominated by different SSU rRNA gene lineages (Figure 2).
Relatives of Candidatus Desulforudis audaxviator, an uncultivated bacterium of the phylum Firmicutes (Chivian et al., 2008), formed the most abundant lineage of clones recovered from borehole 1301A fluids collected in 2008 (36% of clones; Figure 2). This lineage was nearly identical (>99% similarity) to environmental gene clone sequences recovered previously from rock chips incubated within borehole 1301A (Orcutt et al., 2011b) and fluids collected from borehole 1026B (Cowen et al., 2003; Figure 3a). In addition, this lineage was also recovered from borehole 1301A fluids collected in 2009 (7% of clones; Figure 3a), but not in 2010 (Figure 2). Also abundant in the 2008 borehole 1301A clone library was a lineage within the candidate bacterial phylum OP8 (22% of clones; Figure 3b). This phylum was initially identified from a hydrothermal spring within Yellowstone National Park (Hugenholtz et al., 1998) and has since been detected in the marine subsurface (for example, Dhillon et al., 2003; Huber et al., 2006). Borehole 1301A gene clones related to the candidate phylum OP8 identified in this study formed a monophyletic lineage with clones derived from rock chips incubated within borehole 1301A (Orcutt et al., 2011b) and hydrothermal sediments of Guaymas Basin (Teske et al., 2002; Figure 3b). This lineage was also recovered from borehole 1301A fluids collected in 2009 (1% of clones; Figure 3b), but not 2010 (Figure 2). Other less abundant clone groups recovered from borehole 1301A fluid collected in 2008 include members of both the archaeal phyla and the bacterial phyla Actinobacteria, Planctomycetes, Synergistetes, Proteobacteria, Cyanobacteria, and the candidate phyla EM19 and SAR406/marine Group A (Supplementary Table S1 and Supplementary Figures S3 and S4).
Phylogenetic relationships of borehole 1301A fluid-derived SSU rRNA gene clones related to (a) Candidatus Desulforudis audaxviator of the phylum Firmicutes and (b) candidate phylum OP8. Open circles indicate nodes with bootstrap support between 50–80%, whereas closed circles indicate bootstrap support >80%, from 1000 replicates. Gene clones recovered in this study are highlighted in bold font; the relative abundance of identical clones recovered from the same sampling year is listed in parentheses. Gene clones that were targeted for fluorescence in situ hybridization using oligonucleotide probe 1026B3_590 are included within the ‘1026B3 lineage’ bracket. Cultivated Firmicutes (a) and Thermoanaerobacter (b) were used as outgroups (data not shown). The scale bars correspond to 0.1 substitutions per nucleotide position. Relevant gene clones of short length were added after tree construction and bootstrapping, and are indicated by dashed lines.
In contrast to the 2008 sample, a specific lineage within the archaeal Miscellaneous Crenarchaeotic Group (MCG; Takai et al., 2001; Inagaki et al., 2003a), referred to here as the 1026B52 lineage, was the most abundant group of clones recovered from borehole 1301A fluids collected in 2009 (20% of clones; Figure 2). The 1026B52 lineage included a group of clones recovered previously from borehole 1026B fluids (Cowen et al., 2003), as well as clones recovered from borehole 1301A fluids collected in 2008 and 2010 (Figure 4a). In addition to the highly abundant 1026B52 lineage, several less abundant lineages within the MCG were recovered in all three sampling years (Figure 4a). Also abundant in the 2009 borehole 1301A clone library was a lineage related to the genus Desulfocapsa within the Deltaproteobacteria (10% of clones; Figure 2). Closely related clones were also recovered from borehole 1301A fluids collected in 2010, as well as gene clones recovered from such environments as an anaerobic methane-oxidizing sulfate-reducing enrichment (Jagersma et al., 2009), marine sediments of a hydrocarbon seep (Lloyd et al., 2010) and a mud volcano (Pachiadaki et al., 2010) (Figure 4b). Other less abundant clone groups recovered from borehole 1301A fluid collected in 2009 include members of both the archaeal phyla and the bacterial phyla Actinobacteria, Bacteroidetes, Firmicutes, Fusibacter, Verrucomicrobia, Proteobacteria, Cyanobacteria, and the candidate phyla RF3 and OP8 (Supplementary Table S1 and Supplementary Figures S3 and S4).
Phylogenetic relationships of borehole 1301A fluid-derived SSU rRNA gene clones related to (a) the Miscellaneous Crenarchaeotic Group and (b) Desulfocapsa of the Deltaproteobacteria. Cultivated Thermoprotei (a) and Desulfobulbus (b) were used as outgroups (data not shown). Other information as in Figure 3.
The most abundant gene clone group identified from borehole 1301A fluids collected in 2010 was phylogenetically affiliated with a lineage of uncultivated Gammaproteobacteria (27% of clones; Figure 2) and were most closely related to environmental gene clones recovered from hydrothermal sediments (Orcutt et al., 2010; Pachiadaki et al., 2010) and basaltic ocean crust (Santelli et al., 2008; Figure 5a). Closely related clones were also recovered from borehole 1301A fluids collected in 2009 (Figures 2 and 5a). Also abundant in the 2010 borehole 1301A clone library was a lineage related to the family Hyphomicrobiaceae of the Alphaproteobacteria (16% of clones; Figure 2). Close relatives of this gene clone lineage have been recovered from hydrothermal vent chimneys (Suzuki et al., 2004; Voordeckers et al., 2008; Figure 5b). Other less abundant clone groups recovered from borehole 1301A fluid collected in 2010 include members of the archaeal phylum Crenarchaeota and the bacterial phyla Bacteroidetes, Fusibacter, Planctomycetes, Verrucomicrobia, Proteobacteria, and the candidate phylum RF3 (Supplementary Table S1 and Supplementary Figures S3 and S4).
Phylogenetic relationships of borehole 1301A fluid-derived SSU rRNA gene clones related to (a) the JTB35 lineage of Gammaproteobacteria and (b) the Hyphomicrobiaceae lineage of Alphaproteobacteria. Cultivated Methylobacter sp. (a) and distantly related Hyphomicrobiaceae (b) were used as outgroups (data not shown). Gene clones that were targeted for fluorescence in situ hybridization using oligonucleotide probe 1301A10_105_986 are included within the ‘1301A10_105 lineage’ bracket. Other information as in Figure 3.
The three borehole 1301A fluid environmental SSU rRNA gene clone libraries shared four clone groups in common (Supplementary Table S1). Three of these groups were most closely related to microorganisms harboring physiological attributes consistent with the physical and chemical conditions of this environment or related to SSU rRNA gene sequences previously recovered from similar environments, and included the archaeal MCG lineage, the genus Sulfurimonas within the Epsilonproteobacteria, and the genus Thiomicrospira within the Gammaproteobacteria. Within the genus Sulfurimonas, environmental gene clones derived from borehole 1301A fluids did not form a monophyletic lineage, but were all closely related to environmental gene clones derived from a variety of sediment, cold seep and oceanic crustal environments (for example, Li et al., 1999; Huber et al., 2006; Sudek et al., 2009; Supplementary Figure S5a). Within the genus Thiomicrospira, environmental gene clones derived from borehole 1301A fluids formed a monophyletic lineage that was closely related to environmental gene clones originating from hydrothermal fluids (Huber et al., 2006; Kato et al., 2009; Supplementary Figure S5b). The fourth clone group shared by all the three borehole 1301A fluid clone libraries was phylogenetically affiliated with the ubiquitous SAR11 clade of planktonic marine Alphaproteobacteria (Morris et al., 2002). Environmental SSU rRNA gene clone libraries from borehole 1301A fluids collected in 2008 and 2010 did not share any clone groups in common that were also not shared with the 2009 sample.
Direct cell counts and CARD-FISH
After staining with the DNA-specific dye DAPI (4',6-diamidino-2-phenylindole), microscopic examination of borehole 1301A fluid samples revealed average microbial cell abundances of 8.98 × 103 (2009) and 1.53 × 104 (2010) cells ml−1 (Table 3). These values ranged from 9.5 (2009) to 20.1% (2010) of the microbial cell abundance measured in identically processed bottom seawater samples (Table 3). The majority of cells within borehole 1301A fluids appeared as single, free-living cells that were not clumped or otherwise associated with particles (Supplementary Figure S6 and data not shown). However, dense clusters of cells that appeared attached to small (5–10 μm in diameter) particles were occasionally observed (data not shown). Taxon-specific enumeration via catalyzed reporter deposition-fluorescence in situ hybridization (CARD-FISH) revealed Bacteria and Archaea to each make up ca. 30% of the total microbial cells (as determined by DAPI) in borehole 1301A fluids collected in 2009 (Table 3). In 2010, bacterial cells made up nearly 41% of the total counts, whereas archaeal cells accounted for 12% (Table 3). Archaeal cells were predominantly ca. 1 μm diameter cocci, whereas bacterial cells were of diverse morphology (Supplementary Figure S6).
Two clone groups found to be abundant in borehole 1301A fluid libraries were specifically targeted for enumeration via CARD-FISH (Supplementary Table S5). The gene clone lineage related to Candidatus Desulforudis audaxviator of the bacterial phylum Firmicutes made up 32% of the bacterial cells in borehole 1301A fluids collected in 2009. Consistent with the SSU rRNA gene clone library findings, this lineage was below the limit of detection (276±79 cells ml−1) in borehole 1301A fluid collected in 2010. Cells hybridized with the Candidatus Desulforudis audaxviator-specific oligonucleotide probe were ca. 1 × 5 μm rods (Supplementary Figure S6). The gene clone lineage related to an uncultivated group, JTB35, of the Gammaproteobacteria made up 12% of the bacterial cells in borehole 1301A fluids collected in 2009, and 9% of the bacterial cells in fluids collected in 2010 (Table 3). Cells hybridized with the JTB35-related oligonucleotide probe specific to the borehole clones retrieved in this study were ca. 1–2 μm long and ovoid in shape (Supplementary Figure S6). The Candidatus Desulforudis audaxviator and JTB35 gene clone lineages were not detected in bottom seawater samples collected in either 2009 or 2010.
Discussion
Few direct measurements are available to constrain the magnitude of microbial biomass residing within basalt-hosted deep subseafloor basement aquifers (Cowen et al., 2003; Huber et al., 2006). This study is the first to utilize a second-generation CORK-II equipped with a stainless steel fluid delivery line that, after flushing, yielded uncontaminated ocean crustal fluid samples derived directly from the crustal aquifer. The chemical signature of borehole 1301A fluids (Lin et al., 2012) is consistent with previous results from basement fluids collected from borehole 1026B (Wheat et al., 2000; Cowen et al., 2003) and the Baby Bare ridge flank outcrops (Mottl et al., 1998; Sansone et al., 1998; Wheat et al., 2000). In this case, efforts to ensure sampling of pristine crustal fluids produced cellular abundances from borehole 1301A that are 83–93% less than previous reports from borehole 1026B (Cowen et al., 2003; Huber et al., 2006) and 59–67% less than Baby Bare Seamount-derived crustal fluid (Huber et al., 2006). In addition to the lower cell counts, flocs and clumps of cells noted previously in borehole 1026B fluids (Cowen et al., 2003; Huber et al., 2006) were rarely observed in the borehole 1301A fluid samples. Differences in cellular abundance measurements between the two boreholes are most readily explained by differences in CORK design: the CORK affixed to borehole 1301A greatly reduces the possibility of fluid interaction with casing cement and steel liners (Fisher et al., 2005b; Wheat et al., 2011), and the potential for sampling biofilms attached to these materials. However, because the borehole 1301A and Baby Bare Seamount samples used for direct cell counts yielded floc-free fluid with chemical signatures, indicating that they were high-integrity crustal fluids, the differences in cellular abundance may reflect bona fide differences between the two sites. This is further supported by differences in microbial community structure, as microorganisms prevalent in seawater made up a large fraction of the Baby Bare fluid-derived microbial community (Huber et al., 2006), but were rare in fluids derived from borehole 1301A. Thus, this study provides robust data that constrains the magnitude of microbial communities residing in the hydrothermal fluids of the deep-ocean crustal biosphere, albeit at a lower quantity than previous estimates.
Unexpectedly, significant differences in microbial community structure were observed over three consecutive years of sampling crustal fluids from borehole 1301A. Between years, overall community structure was statistically different, the most abundant microbial lineages changed, and absolute cellular abundances of both Bacteria and Archaea varied. This suggests a temporally dynamic microbial community. Borehole 1301A was drilled, cased and equipped with a CORK during the summer of 2004. From the time of drilling until September 2007, bottom seawater flowed into the borehole, resulting in fluids that possessed chemical characteristics identical to the surrounding seawater environment (Wheat et al., 2010). Abruptly (that is, within 1 week), fluid flow reversed, resulting in the production of warm fluids of a chemical composition consistent with the high-integrity basement fluids (Wheat et al., 2010). It is feasible, perhaps even likely, that successional changes in basement fluid microbial community structure in response to such an abrupt change in chemistry are much more gradual, resulting in the inter-annual differences observed here. The observed temporal variation is, perhaps, not unlike the temporal succession of microbial communities thought to occur during the active-to-inactive transition of hydrothermal chimneys and associated venting fluids (for example, Hentscher and Bach, 2012; Sylvan et al., 2012). However, it is also feasible that the variability in community structure is unrelated to the specific turnaround event experienced in September 2007. Instead, it may reflect not well-understood patchiness in the physical and/or chemical characteristics of this system, or result from changes in our methodological approach from year to year (Lin et al., 2012). Continued temporal sampling should help to identify and differentiate stochastic and deterministic processes influencing the structure of microbial communities inhabiting borehole 1301A fluids.
Despite inter-annual variability, the microorganisms found to be abundant in borehole 1301A SSU rRNA gene surveys bared close phylogenetic relationships with cultivated anaerobic thermophiles and/or environmental gene clones previously recovered from related environments, indicating that they are plausible inhabitants of the deep subsurface environment. Environmental gene clones baring close phylogenetic relationships with clones or isolates of apparent seawater origin were infrequent and made up only a small fraction of the libraries originating from the borehole 1301A fluids. One abundant lineage in the borehole 1301A fluid samples from 2008 and 2009 (though curiously not detected by SSU rRNA gene clone library analysis or CARD-FISH in 2010) was nearly identical to the most abundant clone lineage recovered in fluid samples retrieved from borehole 1026B in 2000 (Cowen et al., 2003). This marine crustal fluid lineage forms a monophyletic clade with Candidatus Desulforudis audaxviator and a variety of other environmental gene clones of terrestrial (Moser et al., 2005; Lin et al., 2006; Chivian et al., 2008) and marine (Gihring et al., 2006; Nakagawa et al., 2006; Niemann et al., 2006; Pachiadaki et al., 2010; Orcutt et al., 2011b) deep subsurface origin. Candidatus Desulforudis audaxviator is the tentative name given to the uncultivated bacterium possessing a composite genome constructed from metagenomic DNA sequence data of South African gold mine fracture water origin (Chivian et al., 2008). The 2.35 Mbp genome revealed a motile, sporulating thermophilic chemolithoautotroph capable of sulfate reduction, nitrogen fixation and carbon fixation via the acetyl coenzyme-A synthesis pathway (Chivian et al., 2008), which makes it well suited for an independent lifestyle within the deep subsurface crustal environment. In future studies, it will be valuable to determine the extent to which these metabolic features are shared between abundant members of these distinct but globally related environments, using whole-genome sequence data from the subseafloor-dwelling relatives of Candidatus Desulforudis audaxviator.
The relative percentage of archaeal cells detected within the borehole 1301A fluids was significantly higher than had been measured previously from borehole 1026B and Baby Bear Seamount (12–30% vs <2%; Huber et al., 2006). Although this may partially be explained by improvements in methodology (that is, FISH vs CARD-FISH), analysis via SSU rRNA gene clone libraries supports the conclusion that the differences were not simply methodological. Gene clones related to a major lineage of uncultivated Crenarchaeota known as the MCG (Takai et al., 2001; Inagaki et al., 2003a) were recovered from borehole 1301A fluids in all three sampling years, as well as from borehole 1026B fluids sampled in 2000 (Cowen et al., 2003) and other ocean crustal environments (Huber et al., 2006; Nakagawa et al., 2006; Kato et al., 2009; Orcutt et al., 2011b). The MCG is a diverse microbial assemblage that has been recovered from a wide variety of terrestrial and marine habitats (Teske and Sørensen, 2008), and some evidence indicates that it may be heterotrophic (Biddle et al., 2006; Webster et al., 2010). Consistent with this phylogenetic diversity, recent evidence from fosmid gene clones recovered from marine sediments indicated little gene-order consistency with known archaeal genomes and genomic fragments (Li et al., 2012).
In addition to the abundant Candidatus Desulforudis audaxviator and MCG lineages, several other interesting clone groups worthy of subsequent investigation have emerged from this study. Gene clones related to the candidate bacterial phylum OP8 were detected in consecutive years from borehole 1301A fluids, and have previously been detected in crustal fluids from Baby Bare Seamount (Huber et al., 2006) and rock chips incubated within borehole 1301A (Orcutt et al., 2011b). Gene clones related to the genus Sulfurimonas were recovered in all the three borehole 1301A fluid samples characterized here, which is consistent with their recovery from other chemically reducing environments, such as mid-ocean ridge spreading centers (Santelli et al., 2008), crustal fluids (Huber et al., 2006), whale falls (Tringe et al., 2005) and pelagic redoxclines (Grote et al., 2008). Although evolutionary relatedness between the gene clones identified here and the cultivated representatives of the genus Sulfurimonas is distant, characterized strains are capable of chemolithoautotrophic processes, such as sulfur oxidation and thiosulfate oxidation (Inagaki et al., 2003b; Takai et al., 2006). Members of the Thiomicrospira lineage of Gammaproteobacteria were also recovered in all the three borehole 1301A fluid samples, and characterized isolates of this group are also chemolithoautotrophic sulfur oxidizers (for example, Jannasch et al., 1985; Knittel et al., 2005).
Although we identified a number of microbial lineages that appear to be common inhabitants of the subseafloor crustal fluid environment, none appear to fit within the Ocean Crustal Clade lineages defined previously (Mason et al., 2007, 2009). As the nature of these studies differed (basalt surface-attached vs basalt-hosted fluids), it is possible that, like the marine water column (for example, DeLong et al., 1993), distinct surface-attached and planktonic microbial communities exist within the basaltic ocean crust.
In this study, we focused on cloning and sequencing of PCR-amplified SSU rRNA genes rather than next-generation, deep sequencing of rRNA gene amplicons (for example, Sogin et al., 2006) because of the increase in sequence length afforded by the more traditional approach. Although a massively parallel sequencing strategy would enable much deeper sampling of individual microbial communities than could routinely be achieved by cloning and sequencing, and thus a more comprehensive assessment of the diversity, evenness and community structure contained within our samples (thousands vs hundreds of sequences; Sogin et al., 2006), limitations to the sequencing technology currently restrict it to a short fragment of the 16S rRNA gene. As little is known regarding the diversity of microorganisms inhabiting the deep subsurface basement biosphere, we chose to maximize the phylogenetic signal we could obtain from representative environmental sequences, so that they could serve as quality reference points for future, potentially high-throughput, studies. In addition, longer representative sequences are useful for the identification of taxon-specific regions of variability that can be targeted by oligonucleotide probes in whole-cell in situ assays, such as CARD-FISH, or via quantitative PCR assays targeting extracted nucleic acids. In all 3 years, estimations of richness and rarefaction analysis reveal that there is more microbial diversity to be discovered if deeper sequencing were performed.
The study presented here results from continually evolving sampling equipment and methodology, which serve as a source of variability that may impact our findings or their interpretation. Over the course of the study, the pumping system became better sealed from intruding seawater, permitting a steady increase in the borehole fluid temperature at which sampling began, and an increase in the volume of high-integrity borehole fluid samples. Instead of using a multi-day in situ filtration (Cowen et al., 2003), a clean pump was used here to fill large-volume sampling bags over the course of hours; this relatively rapid sampling method appears to have minimized the contaminating effects of surrounding seawater as evident by the collection of samples that appear to be of pristine ocean basement crustal fluid (Cowen et al., 2012; Lin et al., 2012). The evolution of cleaner sampling equipment and methodology are positive developments for the field of research and represent the best window available into the deep subsurface oceanic crust.
References
Bach W, Edwards KJ . (2003). Iron and sulfide oxidation within the basaltic ocean crust: implications for chemolithoautotrophic microbial biomass production. Geochim Cosmochim Ac 67: 3871–3887.
Baross JA, Wilcock WSD, Kelley DS, DeLong EF, Cary SC . (2004). The subsurface biosphere at mid-ocean ridges: issues and challenges. In: Wilcock WSD, DeLong EF, Kelley DS, Baross JA, Cary SC, (eds). The Subseafloor Biosphere at Mid-Ocean Ridges. American Geophysical Union: Washington, DC, pp 1–11.
Biddle JF, Lipp JS, Lever MA, Lloyd KG, Sørensen KB, Anderson R et al (2006). Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc Natl Acad Sci USA 103: 3846–3851.
Chivian D, Brodie EL, Alm EJ, Culley DE, Dehal PS, DeSantis TZ et al (2008). Environmental genomics reveals a single-species ecosystem deep within earth. Science 322: 275–278.
Cowen JP . (2004). The microbial biosphere of sediment-buried oceanic basement. Res Microbiol 155: 497–506.
Cowen JP, Copson DA, Jolly J, Hsieh C-C, Lin H-T, Glazer BT et al (2012). Advanced instrument system for real-time and time-series microbial geochemical sampling of the deep (basaltic) crustal biosphere. Deep-Sea Res Pt I 61: 43–56.
Cowen JP, Giovannoni SJ, Kenig F, Johnson HP, Butterfield D, Rappé MS et al (2003). Fluids from aging ocean crust that support microbial life. Science 299: 120–123.
Davis EE, Becker K . (2001). Using ODP boreholes for studying sub-seafloor hydrogeology: results from the first decade of CORK observations. Geosci Can 28: 171–178.
Davis EE, Becker K, Pettigrew T, Carson B, MacDonald R . (1992). CORK: a hydrologic seal and downhole observatory for deep-ocean boreholes. Proc ODP Init Repts 139: 43–53.
DeLong EF, Franks DG, Alldredge AL . (1993). Phylogenetic diversity of aggregate-attached vs free-living marine bacterial assemblages. Limnol Oceanogr 38: 924–934.
Dhillon A, Teske A, Dillon J, Stahl DA, Sogin ML . (2003). Molecular characterization of sulfate-reducing bacteria in the Guaymas Basin. Appl Environ Microbiol 69: 2765–2772.
Edwards KJ, Wheat CG, Sylvan JB . (2011). Under the sea: microbial life in volcanic oceanic crust. Nat Rev Microbiol 9: 703–712.
Embley RW, Hobart MA, Anderson RN, Abbott D . (1983). Anomalous heat-flow in the Northwest Atlantic: a case for continued hydrothermal circulation in 80-m.y. crust. J Geophys Res 88: 1067–1074.
Fisher AT, Davis EE, Hutnak M, Spiess V, Zühlsdorff L, Cherkaoui A et al (2003). Hydrothermal recharge and discharge across 50 km guided by seamounts on a young ridge flank. Nature 421: 618–621.
Fisher AT, Urabe T, Klaus A, Wheat CG, Becker K, Davis E et al (2005a). IODP expedition 301 installs three borehole crustal observatories, prepares for three-dimensional, cross-hole experiments in the Northeastern Pacific Ocean. Sci Dril 1: 6–11.
Fisher AT, Wheat CG, Becker K, Davis EE, Jannasch H, Schroeder D et al (2005b). Scientific and technical design and deployment of long-term subseafloor observatories for hydrogeologic and related experiments, IODP Expedition 301, eastern flank of Juan de Fuca Ridge. In: Fisher AT, Urabe T, Klaus A, and the Expedition 301 Scientists. Proceedings of the Integrated Ocean Drilling Program. Integrated Ocean Drilling Program Management International, Inc., 301: College Station, TX doi:10.2204/iodp.proc.301.103.2005.
Fisk MR, Giovannoni SJ, Thorseth IH . (1998). Alteration of oceanic volcanic glass: textural evidence of microbial activity. Science 281: 978–980.
Gihring TM, Moser DP, Lin L-H, Davidson M, Onstott TC, Morgan L et al (2006). The distribution of microbial taxa in the subsurface water of the Kalahari Shield, South Africa. Geomicrobiol J 23: 415–430.
Gold T . (1992). The deep, hot biosphere. Proc Natl Acad Sci USA 89: 6045–6049.
Grote J, Jost G, Labrenz M, Herndl GJ, Jürgens K . (2008). Epsilonproteobacteria represent the major portion of chemoautotrophic bacteria in sulfidic waters of pelagic redoxclines of the Baltic and Black Seas. Appl Environ Microbiol 74: 7546–7551.
Hentscher M, Bach W . (2012). Geochemically induced shifts in catabolic energy yields explain past ecological changes of diffuse vents in the East Pacific Rise 9 degrees 50'N area. Geochem Trans 13: 2.
Huber JA, Johnson HP, Butterfield DA, Baross JA . (2006). Microbial life in ridge flank crustal fluids. Environ Microbiol 8: 88–99.
Hugenholtz P, Pitulle C, Hershberger KL, Pace NR . (1998). Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol 180: 366–376.
Inagaki F, Suzuki M, Takai K, Oida H, Sakamoto T, Aoki K et al (2003a). Microbial communities associated with geological horizons in coastal subseafloor sediments from the Sea of Okhotsk. Appl Environ Microbiol 69: 7224–7235.
Inagaki F, Takai K, Hideki KI, Nealson KH, Horikishi K . (2003b). Sulfurimonas autotrophica gen. nov., sp. nov., a novel sulfur-oxidizing Epsilon-proteobacterium isolated from hydrothermal sediments in the Mid-Okinawa Trough. Int J Syst Evol Microbiol 53: 1801–1805.
Jagersma GC, Meulepas RJ, Heikamp-de Jong I, Gieteling J, Klimiuk A, Schouten S et al (2009). Microbial diversity and community structure of a highly active anaerobic methane-oxidizing sulfate-reducing enrichment. Environ Microbiol 11: 3223–3232.
Jannasch HW, Wirsen CO, Nelson DC, Robertson LA . (1985). Thiomicrospira crunogena sp. nov., a colorless, sulfur-oxidizing bacterium from a deep-sea hydrothermal vent. Int J Syst Bacteriol 35: 422–424.
Kato S, Hara K, Kasai H, Teramura T, Sunamura M, Ishibashi J et al (2009). Spatial distribution, diversity and composition of bacterial communities in sub-seafloor fluids at a deep-sea hydrothermal field of the Suiyo Seamount. Deep-Sea Res Pt I 56: 1844–1855.
Knittel K, Kuever J, Meyerdierks A, Meinke R, Amann R, Brinkhoff T . (2005). Thiomicrospira arctica sp. nov. and Thiomicrospira psychrophila sp. nov., psychrophilic, obligately chemolithoautotrophic, sulfur-oxidizing bacteria isolated from marine Arctic sediments. Int J Syst Evol Microbiol 55: 781–786.
Li L, Kato C, Horikoshi K . (1999). Microbial diversity in sediments collected from the deepest cold-seep area, the Japan Trench. Mar Biotechnol 1: 391–400.
Li PY, Xie BB, Zhang XY, Qin QL, Dang HY, Wang XM et al (2012). Genetic structure of three fosmid-fragments encoding 16S rRNA genes of the Miscellaneous Crenarchaeotic Group (MCG): implications for physiology and evolution of marine sedimentary archaea. Environ Microbiol 14: 467–479.
Lin L-H, Hall J, Onstott TC, Gihring T, Lollar BS, Boice E et al (2006). Planktonic microbial communities associated with fracture-derived groundwater in a deep gold mine of South Africa. Geomicrobiol J 23: 475–497.
Lin H-T, Cowen JP, Olson EJ, Amend JP, Lilley MD . (2012). Inorganic chemistry, gas compositions and dissolved organic carbon in fluids from sedimented young basaltic crust on the Juan de Fuca Ridge flanks. Geochim Cosmochim Ac 85: 213–227.
Lloyd KG, Albert DB, Biddle JF, Chanton JP, Pizarro O, Teske A . (2010). Spatial structure and activity of sedimentary microbial communities underlying a Beggiatoa spp. mat in a Gulf of Mexico hydrocarbon seep. PLoS One 5: e8738.
Mason OU, Di Meo-Savoie CA, Van Nostrand JD, Zhou J, Fisk MR, Giovannoni SJ . (2009). Prokaryotic diversity, distribution, and insights into their role in biogeochemical cycling in marine basalts. ISME J 3: 231–242.
Mason OU, Stingl U, Wilhelm LJ, Moeseneder MM, Di Meo-Savoie CA, Fisk MR et al (2007). The phylogeny of endolithic microbes associated with marine basalts. Environ Microbiol 9: 2539–2550.
Morris RM, Rappé MS, Connon SA, Vergin KL, Siebold WA, Carlson CA et al (2002). SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420: 806–810.
Moser DP, Gihring TM, Brockman FJ, Fredrickson JK, Balkwill DL, Dollhopf ME et al (2005). Desulfotomaculum and Methanobacterium spp. dominate a 4- to 5-kilometer-deep fault. Appl Environ Microbiol 71: 8773–8783.
Mottl MJ, Wheat CG . (1994). Hydrothermal circulation through mid-ocean ridge flanks: fluxes of heat and magnesium. Geochim Cosmochim Ac 58: 2225–2237.
Mottl MJ, Wheat G, Baker E, Becker N, Davis E, Feely R et al (1998). Warm springs discovered on 3.5 Ma oceanic crust, eastern flank of the Juan de Fuca Ridge. Geology 26: 51–54.
Nakagawa S, Inagaki F, Suzuki Y, Steinsbu BO, Lever MA, Takai K et al (2006). Microbial community in black rust exposed to hot ridge flank crustal fluids. Appl Environ Microbiol 72: 6789–6799.
Niemann H, Lösekann T, de Beer D, Elvert M, Nadalig T, Knittel K et al (2006). Novel microbial communities of the Haakon Mosby mud volcano and their role as a methane sink. Nature 443: 854–858.
Orcutt BN, Bach W, Becker K, Fisher AT, Hentscher M, Toner BM et al (2011b). Colonization of subsurface microbial observatories deployed in young ocean crust. ISME J 5: 692–703.
Orcutt BN, Joye SB, Kleindienst S, Knittel K, Ramette A, Reitz A et al (2010). Impact of natural oil and higher hydrocarbons on microbial diversity, distribution, and activity in Gulf of Mexico cold-seep sediments. Deep-Sea Res Pt II 57: 2008–2021.
Orcutt BN, Sylvan JB, Knab NJ, Edwards KJ . (2011a). Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiol Mol Biol R 72: 361–422.
Pachiadaki MG, Lykousis V, Stefanou EG, Kormas KA . (2010). Prokaryotic community structure and diversity in the sediments of an active submarine mud volcano (Kazan mud volcano, East Mediterranean Sea). FEMS Microbiol Ecol 72: 429–444.
Parkes RJ, Cragg BA, Bale SJ, Getliff JM, Goodman K, Rochelle PA et al (1994). Deep bacterial biosphere in Pacific Ocean sediments. Nature 371: 410–413.
Sansone FJ, Mottl MJ, Olson EJ, Wheat CG, Lilley MD . (1998). CO2-depleted fluids from mid-ocean ridge-flank hydrothermal springs. Geochim Cosmochim Ac 62: 2247–2252.
Santelli CM, Orcutt BN, Banning E, Bach W, Moyer CL, Sogin ML et al (2008). Abundance and diversity of microbial life in ocean crust. Nature 453: 653–656.
Schrenk MO, Huber JA, Edwards KJ . (2010). Microbial provinces in the subseafloor. Ann Rev Mar Sci 2: 279–304.
Smith A, Popa R, Fisk M, Nielsen M, Wheat CG, Jannasch HW et al (2011). In situ enrichment of ocean crust microbes on igneous minerals and glasses using an osmotic flow-through device. Geochem Geophy Geosys 12 doi:10.1029/2010GC003424.
Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR et al (2006). Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. Proc Natl Acad Sci USA 103: 12115–12120.
Sudek LA, Templeton AS, Tebo BM, Staudigel H . (2009). Microbial ecology of Fe (hydr)oxide mats and basaltic rock from Vailulu'u Seamount, American Samoa. Geomicrobiol J 26: 581–596.
Suzuki Y, Inagaki F, Takai K, Nealson KH, Horikoshi K . (2004). Microbial diversity in inactive chimney structures from deep-sea hydrothermal systems. Microb Ecol 47: 186–196.
Sylvan JB, Toner BM, Edwards KJ . (2012). Life and death of deep-sea vents: bacterial diversity and ecosystem succession on inactive hydrothermal sulfides. MBio 3: e00279-11.
Takai K, Moser DP, DeFlaun M, Onstott TC, Fredrickson JK . (2001). Archaeal diversity in waters from deep South African gold mines. Appl Environ Microbiol 67: 5750–5760.
Takai K, Suzuki M, Nakagawa S, Miyazaki M, Suzuki Y, Inagaki F et al (2006). Sulfurimonas paralvinellae sp. nov., a novel mesophilic, hydrogen- and sulfur-oxidizing chemolithoautotroph within the Epsilonproteobacteria isolated from a deep-sea hydrothermal vent polychaete nest, reclassification of Thiomicrospira denitrificans as Sulfurimonas denitrificans comb. nov. and emended description of the genus Sulfurimonas. Int J Syst Evol Microbiol 56: 1725–1733.
Teske A, Hinrichs K-U, Edgcomb V, Gomez AD, Kysela D, Sylva SP et al (2002). Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl Environ Microbiol 68: 1994–2007.
Teske A, Sørensen KB . (2008). Uncultured archaea in deep marine subsurface sediments: have we caught them all? ISME J 2: 3–18.
Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW et al (2005). Comparative metagenomics of microbial communities. Science 308: 554–557.
Voordeckers JW, Do MH, Hügler M, Ko V, Sievert SM, Vetriani C . (2008). Culture dependent and independent analyses of 16S rRNA and ATP citrate lyase genes: a comparison of microbial communities from different black smoker chimneys on the Mid-Atlantic Ridge. Extremophiles 12: 627–640.
Webster G, Rinna J, Roussel EG, Fry JC, Weightman AJ, Parkes RJ . (2010). Prokaryotic functional diversity in different biogeochemical depth zones in tidal sediments of the Severn Estuary, UK, revealed by stable-isotope probing. FEMS Microbiol Ecol 72: 179–197.
Wheat CG, Elderfield H, Mottl MJ, Monnin C . (2000). Chemical composition of basement fluids within an oceanic ridge flank: implications for along-strike and across-strike hydrothermal circulation. J Geophys Res-So Ea 105: 13437–13447.
Wheat CG, Jannasch HW, Fisher AT, Becker K, Sharkey J, Hulme S . (2010). Subseafloor seawater-basalt-microbe reactions: continuous sampling of borehole fluids in a ridge flank environment. Geochem Geophy Geosys 11 doi:10.1029/2010GC003057.
Wheat CG, Jannasch HW, Kastner M, Hulme S, Cowen J, Edwards KJ et al (2011). Fluid sampling from oceanic borehole observatories: design and methods for CORK activities (1990–2010). In:Fisher AT, Tsuji T, Petronotis K, (eds). Proceedings of the Integrated Ocean Drilling Program. Integrated Ocean Drilling Program Management International, Inc., 327: College Station, TX doi:10.2204/iodp.proc.327.109.2011.
Acknowledgements
We thank the captain and crew, as well as A Fisher, K Becker, G Wheat and other members of the science teams on board R/V Atlantis cruises ATL15-35, ATL15-51 and ATL15-66, along with the pilots and crew of human-occupied vehicle Alvin and remote-operated vehicle Jason II. We also thank Brian Glazer, Ryan Matsumoto, Michael Matzinger, Michelle Jungbluth, Alberto Robador, Jennifer Murphy, Chih-Chiang Hsieh, Natalie Hamada and Joshua Bninski for sampling and other technical assistance. This research was supported by funding from National Science Foundation Grant MCB06-04014 (to JC and MSR), a Schlanger Ocean Drilling Fellowship (to SPJ), which is part of the NSF-sponsored US Science Support Program for IODP that is administered by the Consortium for Ocean Leadership, the UH NASA Astrobiology Institute and the Center for Microbial Oceanography: Research and Education (C-MORE), a National Science Foundation-funded Science and Technology Center (award no. EF0424599). This study used samples and data provided by the Integrated Ocean Drilling Program. This is SOEST contribution 8695, HIMB contribution 1504 and Center for Deep Biosphere Investigations contribution 134.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Jungbluth, S., Grote, J., Lin, HT. et al. Microbial diversity within basement fluids of the sediment-buried Juan de Fuca Ridge flank. ISME J 7, 161–172 (2013). https://doi.org/10.1038/ismej.2012.73
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2012.73
Keywords
This article is cited by
-
Life strategies for Aminicenantia in subseafloor oceanic crust
The ISME Journal (2023)
-
Decorated Vesicles as Prebiont Systems (a Hypothesis)
Origins of Life and Evolution of Biospheres (2023)
-
Influence of Regional Oceanography and Hydrothermal Activity on Protist Diversity and Community Structure in the Okinawa Trough
Microbial Ecology (2020)
-
Carboxydotrophy potential of uncultivated Hydrothermarchaeota from the subseafloor crustal biosphere
The ISME Journal (2019)
-
Carbon fixation and energy metabolisms of a subseafloor olivine biofilm
The ISME Journal (2019)