Abstract
Robust seasonal dynamics in microbial community composition have previously been observed in the English Channel L4 marine observatory. These could be explained either by seasonal changes in the taxa present at the L4 site, or by the continuous modulation of abundance of taxa within a persistent microbial community. To test these competing hypotheses, deep sequencing of 16S rRNA from one randomly selected time point to a depth of 10 729 927 reads was compared with an existing taxonomic survey data covering 6 years. When compared against the 6-year survey of 72 shallow sequenced time points, the deep sequenced time point maintained 95.4% of the combined shallow OTUs. Additionally, on average, 99.75%±0.06 (mean±s.d.) of the operational taxonomic units found in each shallow sequenced sample were also found in the single deep sequenced sample. This suggests that the vast majority of taxa identified in this ecosystem are always present, but just in different proportions that are predictable. Thus observed changes in community composition are actually variations in the relative abundance of taxa, not, as was previously believed, demonstrating extinction and recolonization of taxa in the ecosystem through time.
Similar content being viewed by others
Introduction
Whether variation in microbial community composition across temporal or spatial gradients arises from differences in relative abundances of taxa that are always present, or from differences in community membership, has direct relevance for microbial ecology (Campbell et al., 2011). If microbial surveys of diversity (Sogin et al., 2006; Fuhrman et al., 2008; Fuhrman, 2009; Gilbert et al., 2009; Kirchman et al., 2009, 2010; Caporaso et al., 2011a, 2011b), which demonstrate changes in the compliment of communities over time and space are actually showing fluctuations in the relative abundance of persistent microbial taxa between the rare biosphere (Sogin et al., 2006) and community dominance (Campbell et al., 2011). This concept, known as the seed-bank hypothesis (Lennon and Jones, 2011) has implications for our understanding of ecological resilience and thresholds to change, especially the reversibility of threshold shifts (Groffman et al., 2006). This concept is elegantly described for marine ecosystems in the One Ocean Model for ecosystem biodiversity (O’dor et al., 2009), whereby microbial life over millions of years can diffuse through the whole ocean, creating a potential seed bank for rapid community adaptation to change. This theoretically enables the community to maintain equilibrium with changing environments, so that the community always prevails (O’dor et al., 2009).
The Western English Channel has been shown to maintain a strong seasonal pattern of microbial species diversity and richness (Gilbert et al., 2009, 2011). These investigations have demonstrated that there were a few very persistent microbial taxa (∼12 operational taxonomic units (OTUs) out of ∼8000 were present every month over 6 years) as shown in other ecosystems (Campbell et al., 2011); also the changes in presence/absence and relative abundance of the rest defined the month in which the sample was taken, as has been shown previously for other coastal times series studies (Fuhrman et al., 2006). However, these studies observed a potential paradox, in that richness was inversely proportional to dominance in the community, so that the month with the greatest richness (December) also had the lowest dominance, or greatest evenness (Gilbert et al., 2011). To explore if the observed changes in community structure between seasons resulted from absolute changes in the community composition or from changes in the relative abundance of the same members, a single time point from the community was sequenced to extreme depth of coverage. This was used to test the hypothesis that ‘the Western English Channel maintains a stable microbial community membership, where the relative abundance of specific taxa fluctuates in response to environmental change’. To test this hypothesis, the sequence identity of 10 729 927 16S rRNA V6 read during December 2007 was compared with an average of 11 481 sequences from every month between January 2003 and December 2008.
Materials and methods
As previously reported, monthly samples collected over a 6-year period (January 2003–December 2008) from the L4 English Channel site (Southward et al., 2005) were subject to amplicon pyrosequencing of the V6 hypervariable region (68 base pairs) of the 16S rRNA gene as detailed previously (Gilbert et al., 2009, 2011). Additionally, the amplicon product used to generate the 16S rRNA pyrosequencing data from December 2007 was resequenced on the Illumina GAIIx platform, providing ∼1000-fold deeper coverage. Illumina sequencing was performed using standard library prep reagents, and v2 clustering and v3 SBS kits. The Illumina paired-end reads were filtered to remove any sequences containing adaptors and then quality filtered during paired end merging, by requiring that at least 10 bases on each end overlapped, and that the overlapping bases were exactly identical. Examination of the quality scores before merging revealed >Q20 across 90% of bases in each read.
Several OTU picking strategies were applied in this study to control for the affect of sequencing error across different platforms, and for the affect of OTU identity threshold. First, all 454 sequences were denoised using the QIIME denoiser program (Reeder and Knight, 2010). OTUs were then picked using a multistep process at both 97% identity and 99% identity, and all OTUs with only a single observation (that is, singletons) were excluded from subsequent analyses. The multistep OTU picking process worked as follows. First, OTUs were picked for the Illumina reads using reference-based uclust (Edgar, 2010) against the greengenes database (DeSantis et al., 2006) clustered at the same percent identity as the OTU picking threshold (that is, either 97% or 99%). Sequences that did not match a greengenes reference sequence at greater than or equal to the identity threshold were allowed to form new clusters (that is, an open-reference OTU picking process). The full-length reference sequence (for OTUs defined by a match to a reference sequence) or the centroid sequence (for OTUs defined by a sequence in the Illumina data set that did not cluster to greengenes) was chosen as a representative sequence for each OTU. Next, OTUs were picked for the 454 pyrosequences in the same open-reference OTU picking process, but against the representative sequences from the Illumina OTU picking run. Again, sequences that did not match a reference sequence were allowed to form new clusters. This process was applied at 97% and 99% identity to form the open-reference OTU picking results (OR97 and OR99). The subsets of OTUs defined by greengenes reference sequences were then extracted to form additional sets of OTU picking results at 97% and 99% identity. These comprise the closed-reference OTU picking results (CR97 and CR99), as no ‘novel’ clusters are included.
Open- and closed-reference OTU picking results underwent parallel analyses to confirm that the results were robust to different types of sequencing error that might arise on the two platforms. Closed-reference OTU picking enables the incorporation of a very strict quality filter; if sequences fail to match a known reference sequence at greater than or equal to the identity threshold, the sequence is excluded from subsequent analyses. All results presented in Figure 1 are based on the open-reference OTU picking process at 97% identity (OR97). These results, as well as the results from the 99% open-reference run, and the 97% and 99% closed-reference runs, are presented as Supplementary Tables 1 and 2, respectively.
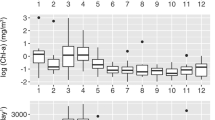
Microbial diversity in the Western English Channel L4 monitoring station: (a) PCoA of unweighted UniFrac. (b) Alpha rarefaction showing phylogenetic diversity summarized by month. (c) Relative abundance of phylum-level taxa over the 6 year time series. The final sample (I1) is abundance in the Illumina sample from December 2007.
Taxonomy summaries are based on the CR97 OTU picking results only, and assigned based on the greengenes sequence defining each OTU. Taxonomy summaries were not generated for the open-reference OTU picking results to avoid artifacts that might arise from assigning taxonomy using the same method (for example, the RDP classifier) on sequences with very different read lengths.
Mean and s.d. sequence counts for the 454 samples and sequence count for the Illumina sample for each OTU picking pipeline are as follows: OR97 454: 11 481±5435, Illumina: 10 729 927; OR99 454: 11 422±5408, Illumina: 10 644 302; CR97 454: 1889±1566, Illumina: 4919 626; CR99 454: 1434±1605; Illumina: 8419 687.
Results
A total of 13 424 OTUs were identified in the combined 454 dataset (including all clustered sequences from all 72 samples), of which 6094 (45%) were singletons (an OTU represented by only a single sequence). These singletons were discarded from the analysis leaving a total of 7330 OTUs for the 454 samples. A total of 116 107 OTUs were identified for the single Illumina sample, of which 56 805 (48%) were singletons. These were discarded leaving a total of 59 302 OTUs for further analysis. Hence, the two sequencing platforms when analyzed using the same pipeline generated similar numbers of singleton OTUs.
Community dissimilarity, measured by unweighted UniFrac distances between all pairs of samples, varied with season (Figure 1a). A similar seasonal pattern was observed in the relative alpha diversities of the samples: winter months have higher within sample diversities than summer months when measured as phylogenetic diversity (Figure 1b) or OTU count (Gilbert et al., 2011). By computing the gain in diversity of each shallowly sequenced 454 samples with respect to the single deeply sequenced Illumina sample, it was possible to assess whether seasonal differences include changes in community membership (in which case, members from the shallowly sequenced summer points would not be present in the deeply sequenced December 2007 sample). When all the pyrosequenced OTUs from the 72 samples were combined into one file, 95.47% of them were found in the single Illumina sample. Additionally, when each of the pyrosequenced samples was compared individually with the Illumina sample, on average 99.75%±0.06 (mean±s.d.) of the OTUs found in the shallow sample were also found in the single deep sample from December 2007 (Supplementary Table S1). Deeper sequencing of any one sample will probably reduce dissimilarity further. The combined OTU diversity from all the 72 shallow 454 samples accounted for 4.64% of the OTUs identified in the single deep Illumina sequenced sample. It is possible that the 95.46% of the Illumina sample OTUs not found in the 454 data could be partially accountable to sequencing error on the Illumina platform, which is starting to be properly understood (Nakamura et al., 2011). However, given that no singletons (OTUs defined by only a single sequence) were included in this analysis, and that the error rate of Illumina sequencing is typically <1% (hence <1 bp error per 68bp sequence, Rodrigue et al., 2010), it is likely that there remains a considerable number of extremely rare bacterial taxa undiscovered in this ecosystem. Strikingly, even with >10 million 16S rRNA fragments sequenced no asymptote was reached on the rarefaction curve for alpha diversity (Figure 1b), suggesting that the taxonomic seed bank for the English Channel L4 site could be vast.
Supplementary Tables S1 and S2 suggest that the apparent difference in alpha diversity by season (Figure 1b) should be interpreted as an observation about the relative evenness of the taxa, rather than an observation about the richness of each community (Gilbert et al., 2011). Additionally, deep sequencing of December 2007, identified a Vibrio taxon, which was previously shown to be usually rare, but bloomed to represent 52% of the pyrosequencing-derived community abundance during August 2003 (Gilbert et al., 2011). In the deep sequenced sample this Vibrio OTU represented 0.029% (320 824 reads) of the reads. In contrast the most abundant taxon, a Rhodobacteriaciae OTU, comprised 13.8 % (1.48 M reads) of the community. Obviously, these abundances are misleading because of differences in 16S rRNA copy number in different bacterial genomes (Klappenbach et al., 2001), and hence these ‘abundances’ are only representative of potential relative abundances. The Illumina and 454 samples from the same date (12 December 2007) exhibit a large apparent difference in their taxonomic composition (Figure 1c) despite clustering close to one another in the UniFrac PCoA plot (Figure 1a). This discrepancy represents a difference in the percent of taxonomically unclassifiable sequences between the two samples: these could appear to be large changes in a taxonomic summary, but if there were a short branch length between the classifiable and unclassifiable sequences they will make a small contribution to the UniFrac distance between the samples.
Discussion and conclusions
Deep sequencing of a single time point from the L4 6 year time series supports the hypothesis that observed seasonal differences in the microbial community composition actually represent fluctuations in community member abundance rather than fluctuations in community membership. In the summer months, some high-abundance taxa may dominate the samples, making richness appear lower when relatively few 16S rRNA amplicons are sequenced. In the winter months, a more even abundance distribution allows more taxa to be sampled when the same number of sequences is examined, causing the richness to appear higher.
Deep sequencing presents a risk in identifying contaminants derived from the physical water sampling or DNA extraction and PCR amplification in the laboratory. These potential contaminants are, however, likely to be very low in abundance and therefore missed in a conventional shallow survey. Although controlling for this contamination is exceptionally difficult, this does not necessarily detract from the observed results: 99.96% of the microbial diversity in the 72 time points can be found in the top 5 % of the abundant OTUs in the deep sequencing time point, suggesting that the 72 time points represented a substantially shallow sequencing of the ecosystem. If these were the results of potential carry over between sampling events 6 months apart, their relative abundances might be expected to be exceptionally lower in this deep sequenced sample. Additionally, whereas the short read length impedes taxonomic identification, no obvious laboratory derived contaminants were observed in this dataset.
The robust cycling of bacterioplankton diversity at L4 (Gilbert et al., 2009, 2011) raises the question of whether unique water masses contain characteristic microbial communities, and hence the observed patterns result from turnover of these water masses. However, this is unlikely, as hydrographic patterns using drift bottles have demonstrated a strong west to east flow through the English Channel (Southward et al., 2005), with the residence of a water mass at the L4 site being ∼2 weeks (Lewis and Allen, 2009). Therefore, the observed cycling of the bacterial community is unlikely to be because of patterns of recolonization of the ecosystem by a microbiome associated with a periodic water mass intrusion, such as from advection of the Celtic Sea in to the Western English Channel, which largely depends on sporadic wind conditions. This study focused on a specific location, and it is not known how representative the observed annual patterns are of the entire English Channel.
The hydrographic patterns associated with the movement of water masses can probably be discounted as evidence for the robust community cyclicity. However, the influence of terrestrial input by river runoff can be substantial. It is therefore possible, that if patterns of increased rainfall, and hence riverine output, were correlated to the seasonal microbial turnover, then it could be considered as a potential explanatory variable. However, riverine input and weather patterns are also sporadic in this region, and hence the intrusion of such water masses into the L4 site, as defined by changes in the salinity and nutrient availability, is not predictable (Gilbert et al., 2011). Although these episodic events do occur they are not correlated to any significant changes in the microbial community diversity at L4 and do not appear to influence the overall stability of the community turnover and seasonal cycle (Gilbert et al., 2011), hence they do not influence a potential threshold change in the ecosystem. Given the lack of evidence for a potential repetitive hydrographic or meteorological event that could produce the remarkable reproducibility of the community profile in each month between years, the other likely scenario is that the same core community is always present in the English Channel, or potentially isolated to the L4 location.
Our results suggest that seasonal differences in marine microbial community profiles over a 6-year time series represent changes in the relative abundances of taxa that are always present. This observation does not negate colonization by ‘alien’ species; it merely indicates that this ecosystem maintains an extremely large core microbiome. Therefore, a disease state or threshold change that involves colonization by a microorganism not normally found in the ecosystem cannot be discounted.
Given the feasibility of such a study, it is now possible to explore whether this principle generalizes to other microbial ecosystems such as soils and host-associated environments. These results have the potential to fundamentally alter our perception of microbial community turnover, succession and extinction. Also, understanding that such a vast and viable microbial seed bank does exist, provides the potential for more informed ecosystem management.
References
Campbell BJ, Yu L, Heidelberg JF, Kirchman DL . (2011). Activity of abundant and rare bacteria in a coastal ocean. Proc Natl Acad Sci USA 108: 12776–12781.
Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J et al. (2011a). Moving pictures of the human microbiome. Genome Biol 12: R50.
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ et al. (2011b). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 108 (Suppl 1): 4516–4522.
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072.
Edgar RC . (2010). USEARCH. http://www.drive5.com/usearch.
Fuhrman JA . (2009). Microbial community structure and its functional implications. Nature 459: 193–199.
Fuhrman JA, Hewson I, Schwalbach MS, Steele JA, Brown MV, Naeem S . (2006). Annually reoccurring bacterial communities are predictable from ocean conditions. Proc Natl Acad Sci USA 103: 13104–13109.
Fuhrman JA, Steele JA, Hewson I, Schwalbach MS, Brown MV, Green JL et al. (2008). A latitudinal diversity gradient in planktonic marine bacteria. ProcNatl Acad Sci USA 105: 7774–7778.
Gilbert JA, Field D, Swift P, Newbold L, Oliver A, Smyth T et al. (2009). The seasonal structure of microbial communities in the Western English Channel. Environ Microbiol 11: 3132–3139.
Gilbert JA, Steele JA, Caporaso JG, Steinbrück L, Reeder J, Temperton B et al. (2011). Defining seasonal marine microbial community dynamics. ISME J; e-pub ahead of print 18 August 2011; doi:10.1038/ismej.2011.107.
Groffman P, Baron J, Blett T, Gold A, Goodman I, Gunderson L et al. (2006). Ecological thresholds: the key to successful environmental management or an important concept with no practical application? Ecosystems 9: 1–13.
Kirchman DL, Campbell BJ, Yu L, Straza TRA . (2009). Temporal changes in bacterial rRNA and rRNA genies in Delaware (USA) coastal waters. Aquat Microbial Ecol 57: 123–135.
Kirchman DL, Cottrell MT, Lovejoy C . (2010). The structure of bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16S rRNA genes. Environ Microbiol 12: 1132–1143.
Klappenbach JA, Saxman PR, Cole JR, Schmidt TM . (2001). rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res 29: 181–184.
Lennon JT, Jones SE . (2011). Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat Rev Microbiol 9: 119–130.
Lewis K, Allen JI . (2009). Validation of a hydrodynamic-ecosystem model simulation with time-series data collected in the Western English Channel. J Mar Syst 77: 296–311.
Nakamura K, Oshima T, Morimoto T, Ikeda S, Yoshikawa H, Shiwa Y et al. (2011). Sequence-specific error profile of Illumina sequencers. Nucleic Acids Res 39: e90.
O’dor RK, Fennel K, Vanden Berghe E . (2009). A one ocean model of biodiversity. Deep-Sea Res Part Ii: Topical Stud Oceanogr 56: 1816–1823.
Reeder J, Knight R . (2010). Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat Methods 7: 668–669.
Rodrigue S, Materna AC, Timberlake SC, Blackburn MC, Malmstrom RR, Alm EJ et al. (2010). Unlocking short read sequencing for metagenomics. PLoS One 5: e11840.
Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR et al. (2006). Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. Proc Natl Acad SciUSA 103: 12115–12120.
Southward AJ, Langmead O, Hardman-Mountford NJ, Aiken J, Boalch GT, Dando PR et al. (2005). Long-term oceanographic and ecological research in the Western English Channel. AdvMar Biol 47: 1–105.
Acknowledgements
This work was supported by the US Department of Energy under Contract DE-AC02-06CH11357. We acknowledge the support of Plymouth Marine Laboratory and the Western Channel Observatory for access to samples acquired from the L4 observatory in the English Channel.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Supplementary information
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Caporaso, J., Paszkiewicz, K., Field, D. et al. The Western English Channel contains a persistent microbial seed bank. ISME J 6, 1089–1093 (2012). https://doi.org/10.1038/ismej.2011.162
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2011.162
Keywords
This article is cited by
-
Succession of bacterial biofilm communities following removal of chloramine from a full-scale drinking water distribution system
npj Clean Water (2023)
-
Salp blooms drive strong increases in passive carbon export in the Southern Ocean
Nature Communications (2023)
-
Influence of short and long term processes on SAR11 communities in open ocean and coastal systems
ISME Communications (2022)
-
Evaluation of the gut microbiome in association with biological signatures of inflammation in murine polytrauma and shock
Scientific Reports (2021)
-
Significant Bacterial Distance-Decay Relationship in Continuous, Well-Connected Southern Ocean Surface Water
Microbial Ecology (2020)