Abstract
Much of the phylogenetic diversity in microbial systems arises from rare taxa that comprise the long tail of taxon rank distribution curves. This vast diversity presents a challenge to testing hypotheses about the effects of perturbations on microbial community composition because variability of rare taxa among environmental replicates may be sufficiently large that it would require a prohibitive degree of sequencing to discern differences between samples. In this study we used pyrosequencing of 16S rRNA tags to examine the diversity and within-site variability of salt marsh sediment bacteria. Our goal was to determine whether pyrosequencing could produce similar patterns in community composition among replicate environmental samples from the same location. We hypothesized that repeated sampling from the same location would produce different snapshots of the rare community due to incomplete sequencing of the taxonomically rich rare biosphere. We demonstrate that the salt marsh sediments we sampled contain a remarkably diverse array of bacterial taxa and, in contrast to our hypothesis, repeated sampling from within the same site produces reliably similar patterns in bacterial community composition, even among rare organisms. These results demonstrate that deep sequencing of 16s tags is well suited to distinguish site-specific similarities and differences among rare taxa and is a valuable tool for hypothesis testing in microbial ecology.
Similar content being viewed by others
Introduction
For decades microbial ecologists faced the challenge of inferring microbial community composition from modest-sized ribosomal RNA (rRNA) data sets that represented amplicon libraries from environmental DNA. Even large amplicon libraries (>1000 sequences) often represented only a very small fraction of the different taxa present in most source communities although a few studies collected on the order of 70 000 sequences (Ley et al., 2006). As a result, a number of mathematical models have been proposed to extrapolate composition and richness of microbial communities based on relatively small sample sizes (summarized in Lozupone and Knight, 2008 and Schloss, 2008).
As the first next-generation sequencer became commercially available in 2005, massively parallel DNA sequencing protocols such as pyrosequencing have become preferred tools for examining microbial community composition because they allow researchers to sequence more deeply into a community than had previously been possible with the time and cost constraints of Sanger sequencing (Margulies et al., 2005; Sogin et al., 2006). One result of this tremendous advance in sequencing capability is the recognition, for the first time, of the vast diversity of low abundance microbial taxa that exist in surface and deep sea waters (Sogin et al., 2006; Huber et al., 2007), soil (Roesch et al., 2007), and human gut (Turnbaugh et al., 2009) ecosystems. Kunin et al. (2009) argued that much of the diversity described in these initial studies was a result of sequencing error. The error rate of these methods after appropriate quality control procedures, however, is quite low (Huse et al., 2010). Reanalysis of these initial studies with new clustering methods that minimally inflate the number of operational taxonomic units (OTUs) report only slightly lower richness estimates with rank abundance curves that indicate a large abundance of rare taxa (Huse et al., 2010). New research is needed to understand the ecological and evolutionary role of the rare biosphere, although evidence already suggests that these rare organisms do display biogeography (Galand et al., 2009), and that they provide a source pool of diversity that allows microbial communities to respond to environmental change (Brazelton et al., 2010).
The ability to detect how environmental perturbation alters low abundance microbial taxa (defined operationally in this study as sequences present on average less than five times in 20 000–25 000 tag sequences or from 0.02–0.025% of the time), requires that the repeatability of the rare biosphere within a particular site be sufficiently consistent that variation between two different sites can be inferred. If low abundance taxa represent a universal source pool of bacteria (the ‘everything’ in Baas Becking’s (1934) hypothesis ‘everything is everywhere’), we hypothesized that it would be challenging to infer meaningful differences between the rare biospheres of two different samples, even with the depth of sequencing currently possible. However, if there is some sort of environmentally driven functional selection acting on the rare members of the microbial community then, assuming sufficient sampling depth, there should be greater similarity in the rare biospheres of environmental replicates than from samples taken from two different locations.
The logic of this argument is as follows: if the rare biosphere represents a source pool of microbes that results from universal dispersal then repeated samples taken from the same site, when not sequenced to completion, will display a snapshot of the rare taxa that is selected at random from all the low abundance taxa present. Any similarity that happens to exist among the community composition of low abundance taxa in repeated samples would be a result of the chance sequencing of the same equally rare organisms. If this is true, a snapshot of the rare biosphere taken from two environmental replicates should be roughly as dissimilar as a snapshot taken between two different samples because in all cases we are subsampling from the same universal source pool. If, however, everything is not everywhere; if environmental factors, rather than universal dispersal, drive the distribution of microorganisms from the most abundant to the most rare, then replicate sampling from the same location should result in similar snapshots of the microbial community. If pyrosequencing is to be a useful tool for testing ecological hypotheses regarding microbial community compositional shifts along gradients, or that result from disturbance, the repeatability in the rare biosphere among environmental replicates must be sufficiently high that consistent patterns can be distinguished.
In light of these considerations, we assessed the variability of microbial community compositions in replicate environmental samples taken over very small spatial scales in salt marsh sediments. Salt marshes serve critical functions in marine habitats and the phylogenetic diversity of their microbial communities exceeds that of most other environments including species-rich soils (Lozupone and Knight, 2007). Salt marshes have a key role in protecting adjacent coastal habitats from human-derived influence (Valiela and Cole, 2002) and because marshes are precariously located between terrestrial uplands and marine waters, they are vulnerable to environmental perturbations from both environments. Many of the ecosystem services provided by salt marshes are microbially mediated, yet little is known about the extent of diversity in these key habitats. Achieving a comprehensive understanding of the role that this microbial diversity has in ecosystem-scale processes in salt marshes first requires an understanding of whether incomplete sequencing distorts our ability to define the composition of the rare community among environmental replicates.
The objectives of this study are threefold. First, we document the repeatable pattern of bacterial diversity in environmental replicates from one location in salt marsh sediments. In addition to random variation that would result from error associated with DNA extraction, PCR amplification and sequencing misreads (all which can be assessed by examining technical replicates) there is additional potential variation that can result from fine-scale variation found within the environment. This additional variation must be assessed to ascertain whether differences between two unreplicated samples are meaningful when compared with differences among environmental replicates. Second, by examining diversity in both individual and pooled samples taken from the same location in the marsh, we assess the within-site variability in the sediment microbial community. We hypothesize that pooling and homogenizing sediments from a number of samples, and taking a subsample of the pool will decrease within-site variability and lead to more repeatable patterns in community composition because it will minimize patchiness that results from fine-scale environmental variability. Finally, we compare community composition in individual and pooled marsh sediments with community composition in an out-group sample taken from the water column draining an adjacent marsh creek. This nearby community should contain a mix of typical pelagic taxa and marsh sediment taxa that get resuspended from the marsh platform and are transported to coastal waters through the draining marsh creeks. The differences between the sediment and water column communities provides a test of whether the rare biosphere of replicated samples can be consistently differentiated from the rare biosphere of an out-group sample. If the rare biospheres of the replicate samples are considerably more similar to each other than they are to the rare biosphere of the out-group sample, it suggests that functional selection with the marsh sediments is sufficiently strong that the community can be repeatedly deciphered despite incomplete sequencing.
Materials and Methods
Sample collection
We collected samples from the Great Sippewissett Salt Marsh in Falmouth (MA, USA) (41° 34.58 N, 70° 38.23 W) on 10 September 2008 from within a 100 cm2 area of unvegetated marsh sediments within the tall Spartina alterniflora habitat. As the goal of this study was to establish the degree of variability within environmental replicates, we selected our samples so as to minimize environmental variability; thus, all samples were collected from sediments that were approximately equidistant from any S. alterniflora stems and in areas that had the same elevation above mean sea level, so as to avoid any variations in redox chemistry associated with tidal inundation. A sterile 5-ml syringe core was used to sample the top 1 cm of marsh sediment. Six individual samples were taken and extruded immediately into separate 2-ml cryovials that were stored on ice and then transferred to a −80 °C freezer at the Marine Biological Laboratory in Woods Hole (MA, USA). An additional 12 sediment cores were also taken from the same 100 cm2 area; 6 of the 12 cores were pooled in a sterile 20 ml scintillation vial and the remaining six were extruded into a second scintillation vial. These vials were stored on ice and returned to the lab, where they were homogenized with a sterile spatula. Subsamples from each of the pooled and homogenized cores were removed and stored at −80 °C in 2-ml cryovials. The microbial community from the water column of a creek draining the adjacent Little Sippewissett Salt Marsh that was sampled on 10 July 2007 served as an out-group. One litre of water was collected in a triple-rinsed Nalgene bottle and returned on ice to the lab for filtration. The 1-l sample was vacuum filtered through a Sterivex filter, lysis buffer was added, and the filter unit was stored at −80 °C until DNA extraction.
DNA extraction and amplification
DNA from 0.5 g of marsh sediment was extracted using the PowerSoil DNA Isolation kit (MoBio Laboratories, Carlsbad, CA, USA) following manufacturer’s instructions. DNA from the water column sample was extracted using the Gentra PureGene DNA extraction kit (Qiagen, Valencia, CA, USA) also following the manufacturer’s instructions. The hypervariable V6 region of the bacterial 16S rRNA gene was amplified using a cocktail of five forward and four reverse primers that amplify the 16S rRNA genes from the majority of known bacteria (Huber et al., 2007). The primers contain the Roche A and B adapters fused to a 5-nucleotide multiplex identifier and terminated by 19 bp that complement conserved regions flanking the bacterial 16S rRNA genes. The multiplex identifier allows the bioinformatic identification of pyrosequencing reads from multiple samples in a single pyrosequencing analysis (Huber et al., 2007). Amplified DNA was purified using a MinElute PCR Purification kit (Qiagen) and quality and quantity of the DNA was confirmed on a Bioanalyzer 2100 (Agilent, Palo Alto, CA, USA) before sequencing on a Roche GSFLX pyrosequencer. Further details on these methods have been published elsewhere (Sogin et al., 2006; Huber et al., 2007; Huse et al., 2007, 2008, 2010).
Data analysis
After sequencing, data were subjected to rigorous quality control checks as described previously (Huse et al., 2007, 2008, 2010). These quality control measures included the removal of all reads that had any ambiguous base calls, that had read lengths longer than the typical distribution of sequence lengths, or that had inexact matches to the initial primers. With these quality checks in place, the read error rate associated with pyrosequencing was reduced to <0.2% (Huse et al., 2007). Sequences that passed quality checks were trimmed to remove both primers and were then assigned taxonomy using GAST (Global Alignment for Sequence Taxonomy; Huse et al., 2008). The single linkage preclustering algorithm (Huse et al., 2010) used nearest neighboring on rank abundance-sorted sequences to identify 2% preclusters, and average neighboring in mothur (Schloss et al., 2009) to identify 3%, 6% and 10% clusters (OTUs). Huse et al. (2010) demonstrate that OTU inflation resulting from multiple sequence alignment followed by complete linkage clustering can be minimized via the single linkage preclustering pipeline used to analyze these data. Moreover, they indicate that this analysis pipeline, when applied to the short reads sequenced here, reduced OTU inflation without changing the fraction of taxa that comprise the long tail of the taxa distribution curve because it preserves the correct proportion of singletons, doubletons, and tripletons, while eliminating noise by clustering errant sequences with the appropriate parent sequence. Further analysis by Huse et al. (2010), Quince et al. (2011), and Schloss et al. (2011) indicates that treatment of pyrosequencing data by 2% single linkage preclustering produced results that are similar to results produced via PyroNoise with chimera checking (Quince et al., 2009).
After clustering the data using the algorithms described above, we used the CatchAll software program (Bunge et al., 2012) to calculate nonparameteric ACE and Chao1 richness indices. For the remaining analyses, all data were normalized to the sample that contained the highest number of sequence tags (ENV 1: 24 675 (range: 20 783–24 675). We used EstimateS (Version 8.0.0, RK Colwell, http://purl.oclc.org/estimates) to calculate similarity matrices using the Bray Curtis similarity index on the normalized data (CN=2jN/(aN+bN), where aN=total number of individuals in site A, bN=total number of individuals in site B and jN=the sum of the lower of the two abundances in both samples). In R, we used the vegdist program to calculate dissimilarities and we used hclust to construct phenograms using average linkage clustering, which is an Unweighted Pair Group Mean (UPGMA) method of analysis. The cumulative frequency histograms were calculated on natural log transformed abundance data using the GraphPad Software (La Jolla, CA, USA) statistical package Prism. Curve fit parameters were determined in Prism by fitting Gaussian curves to the data using a least squares fit.
Results and discussion
Salt marsh microbial diversity
Of the 42 phyla recognized at the time of these analyses, all but one, Caldiserica, was present at least one time in our salt marsh samples (Supplementary Table S1). Sediments in this region of the marsh were dominated by the Proteobacteria (61.1±2.9%), but had considerable contributions from Bacteroidetes (9.4±2.9%), Acidobacteria (7.0±0.5%), Planctomycetes (4.6±0.8), Verrucomicrobia (4.4±0.9%), Chloroflexi (3.2±0.9%) and Gemmatimonadetes (2.9±0.7%). By contrast, the water column sample used as an out-group was >90% Proteobacteria, with a minor contribution from Bacteroidetes (7%) and Cyanobacteria (1%). The remaining 27 phyla present accounted for <2% of the reads from the water column sample (Supplementary Table S1). We examined the distribution of orders within the Proteobacteria to further describe the community composition of the sediment samples. Within the Proteobacteria there were 47 recognized orders of which 39 were present in the marsh sediment samples (Supplementary Table S2). The most abundant orders were roughly evenly split among Rhodobacterales (12%), Myxococcales (13%), unidentified deltaproteobacteria (10%), and Xanthomonadales (14%). Of these dominant orders, only Rhodobacterales was also numerically important in the water column out-group sample. The other two orders that dominated the water column sample were Rickettsiales, of which the ubiquitous pelagic bacteria SAR11 is a member, and Alteromonadales (Supplementary Table S2).
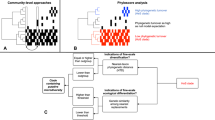
Analysis of samples at the phylum and order levels indicated strong similarity among the sediment samples, and at both levels of biological classification the sediments were quite different than the water column out-group (Supplementary Tables S1 and S2), but the dominant members of the community may drive these conclusions. A higher resolution analysis of the microbial community composition can be performed at the species level (Figure 1). Using the software present in the Visualization and Analysis of Microbial Population Structure analysis pipeline (http://vamps.mbl.edu/), we plotted the relative abundance of bacterial species in each of the sediment samples and in the water column out-group sample taken from Little Sippewissett Marsh (LSM). When all species were included in the analysis (Figure 1a), there were clear similarities among all sediment samples and they were distinctly different than the water column sample although these similarities may, in part, be owing to the relative importance of the dominant members of the community. To get a better look at the distribution of the remaining community composition, we removed the dominant taxa (defined as those accounting for more than 1% of the population) and still saw a greater degree of cohesion among the sediment samples than between the sediment and water samples (Figure 1b). This apparent cohesion in taxonomic identity among the sediment samples suggests that the potentially confounding effects of within-site heterogeneity and incomplete sequencing are not sufficiently strong that repeatable patterns in community composition cannot be discerned.
Stacked bar plots of the bacterial species present in sediment samples and in a water column out-group sample. (a) The relative abundance of all species present and (b) the relative abundance of those species present <1% of the time. There are too many species in each sample to make a legend decipherable but the species data are publically available at www.vamps.mbl.edu. The data include six sediment samples that were collected individually (ENV1–ENV6), two that were subsampled from pooled sediments (HOM1 and HOM2) and one from the water column draining an adjacent marsh.
Next, we used the clustering methodology described by Huse et al. (2010) to calculate rarefaction curves (Figure 2) and different estimators of richness (Table 1) and diversity (Table 2), for each of the eight sediment samples and the water column out-group sample at three different degrees of clustering, 3% (Figure 2a), 6% (Figure 2b) and 10% (Figure 2c). In all cases there was considerable overlap between the slopes of the individual (ENV1–ENV6) and pooled (HOM1 and HOM2) samples, though all sediment samples had steeper slopes than the water column sample, suggesting that there was a considerable amount of unidentified diversity. Furthermore, even at the 10% clustering level the slope of the sediment rarefaction curves remain curvilinear, further evidence that there is likely considerable diversity yet to be sequenced. The rarefaction curves indicate an essential point—we have not begun to approach sequencing these samples to completion, thus although we define ‘rare’ in this case as those sequence present fewer than five times per sample, this is a relative measure. These sequences are rare compared with the highly abundant sequences we uncovered, however, the truly rare sequences that are present only a few times in a gram of sediments likely escape detection owing to incomplete sequencing.
Additional estimators of taxonomic richness, the Chao and ACE estimators were calculated based on tags clustered at 3%, 6% and 10% sequence divergence (Table 1, Supplementary Table S3). Depending on the analysis, these estimators can be sensitive to the depth of sampling, with increased sampling leading to artificial inflation of diversity (Gihring et al., 2012). Following the single linkage preclustering clustering used to analyze these data (Huse et al., 2010), the average OTU inflation resulting from sampling intensity is ∼1–2 OTU for every 1000 sequence reads (Huse et al., 2010). With the samples included here, the yield of high-quality sequences from each sample ranged from 20 783–24 675 sequences. Thus possible richness inflation due to the maximum sampling differences of 3892 sequences among samples was not likely to exceed 8–10 sequences, numbers that fall well within the confidence intervals of the richness estimators (Supplementary Table S3). Thus we take Chao and ACE to be good estimators of taxonomic richness. At the 3% clustering level, each of the sediment samples contained twice as many observed OTUs (∼4100) as did the water column sample (∼1850 OTUs). Chao and ACE estimators tend to underestimate actual richness owing to their extrapolation from small sample sizes (Hong et al., 2006; Quince et al., 2008). However, as a minimum estimate, these estimators indicated that there are between 7000–10 000 bacterial OTUs in the sediments when clustered at 3% sequence divergence (Table 1). This surpasses the Chao estimates of richness for 3% clusters in the water column, but the ACE estimator of bacterial 3% OTUs in the water column sample was roughly equivalent to the sediment sample estimates. When clustered at the 6% and 10% sequence divergence levels, both richness metrics indicated that the estimated taxonomic richness in the water column sample was considerably lower than the estimated richness of the sediment samples (Table 1). We also calculated the Shannon Diversity indices calculated at the standard 3% level of sequence divergence (Table 2), and these data also support our conclusion that there is greater diversity in the sediments than in the water column (Table 2). These estimates of diversity and richness are within the range reported for other soils and sediments (Jørgensen and Boetius, 2007; Roesch et al., 2007; Morales et al., 2009).
It is important to recall that the diversity present in these samples is only representative of the diversity found in a small area of one location in the marsh and cannot be taken to represent the entire diversity present in salt marshes. As such, these data are conservative estimates of diversity as other habitats within the marsh may well harbor additional diversity not uncovered in the small scale sampling area examined here. Several factors may have contributed to the tremendous bacterial diversity found in these salt marsh sediments. Located between terrestrial uplands and marine waters, salt marshes are strongly influenced by both habitats (Valiela and Teal, 1979) and may retain legacies of both microbial source communities. Steep and fluctuating redox gradients in salt marshes (Howes et al., 1981) also suggest a wide range of electron acceptors available to support microbial metabolisms. Different mineral fractions of soils have distinct bacterial communities (Carson et al., 2009), so variations in mineral content of the marsh could increase microbial diversity. Furthermore, organic matter has tremendous spatial complexity at small scales (Lehman et al., 2008), so organic rich salt marsh sediments likely have considerable diversity associated with niche differentiation around organic aggregates.
Comparison of individual and homogenized samples
The factors that promote diversity in marsh sediments also act to promote patchiness within those sediments. We hypothesized that this patchiness, when combined with the stochastic nature of incomplete sequencing, would lead to high variability among environmental replicates that would make it difficult to determine real differences among unreplicated samples. The data, however, demonstrate a degree of similarity in community composition among multiple samples collected from within the same location (Figure 1, Supplementary Tables S1 and S2), suggesting that within-site variability is small, even at the species level. Further evidence that within-site variability is small can be gleaned from a comparison of the individual samples with the pooled samples. We hypothesized that pooling multiple sediment cores and sequencing a subsample from the pooled and homogenized sediments would produce a snapshot of the community that would be more representative than any single snapshot from individual samples. By sequencing the pooled subsample to the same depth as each of the individual samples, the data would be skewed toward those taxa that were present in multiple subsamples. This would decrease the importance of patchy taxa and of the very minor constituents of the rare community that were only present in one or two of the subsamples. The result would be a repeatable assessment of within-site variability, a necessary step for subsequent hypothesis testing.
The community composition in the pooled samples (HOM1 and HOM2) does not appear to be tremendously different from the individual samples (ENV1–ENV6, Figure 1). The only plausible explanation for this similarity is that the community composition of each of the pooled subsamples was roughly similar to each of the individual samples. If there were patches of different microbes that were locally abundant (present in one or two subsamples but not in all six), this would skew the taxa abundances in the homogenized samples such that they would be different than the individual samples. That the data do not demonstrate this skew in either homogenized sample lends further support to the conclusion that the within-site variability in these sediments is small. Creating a mechanism to quantify within-site variability will allow for the identification of a baseline community so that deviations from the baseline can be observed.
Quantifying similarities among samples
If pyrosequencing is to be effectively used to examine differences in microbial communities either along environmental gradients, or that result from environmental perturbations, within-site variability must be quantified sufficiently well that a different site (or a postdisturbance community within the same site) can be distinguished. If sites were entirely dominated by a few numerically abundant taxa that differ from location to location, this would be a relatively simple statistical test. Most pyrosequencing data, however, suggest the presence of a long tail of low abundance taxa that exist in many habitats (for example, Sogin et al., 2006). It is therefore not sufficient to examine differences among dominant taxa; it must also be possible to quantify similarities and differences among the rare members of the microbial community.
As a first step, we quantified the differences in bacterial community composition of the normalized individual and homogenized sediment samples using the Bray–Curtis similarity index (Magurran, 1988). We then calculated similarities between the sediment samples and the water column out-group sample. The input data for these analysis came from the GAST taxon assignments generated via the Marine Biological Laboratory’s VAMPS pipeline (http://vamps.mbl.edu/). We first compared similarities across the entire community of microbes (first column), by normalizing the number of sequences per sample with the number of sequences found in the most abundant sample. We then sorted the data by average abundance across all samples and recalculated the similarity index values just for those samples that had average abundances that fell within each of the bins. Thus, we calculated completely independent similarity matrices for bins that contained the most abundant taxa (operationally defined as those taxa present, on average, more than 100 times), bins that contained only the rare taxa (operationally defined as those present, on average, fewer than 5 times) and the various clusters in between those two extremes (Figure 3).
We hypothesized that environmental selection within sediment samples would lead to considerable similarities among the most abundant taxa but that even among the most abundant taxa the sediments would have little similarity with the water column out-group. Furthermore, when comparing taxa with low abundances, the community similarity in replicate sediment samples would go down because incomplete sequencing would lead to a snapshot of taxa selected at random from all the low abundance taxa present in each sample. We feared that this stochastic element would increase dissimilarity among sediments and would make interpreting results of experimental perturbations difficult. If the dissimilarity created by incomplete sequencing of replicate samples was sufficiently large, there would be as much dissimilarity among the replicate sediment samples as there would be between the sediment samples and the out-group water sample.
We were correct that the abundant taxa in the sediment samples were similar to one another both within the individual environmental replicates (Figure 3, blue columns) and between the individual and homogenized samples (Figure 3, red columns). It was also not surprising that the dominant members of the sediment bacterial community were considerably different than the dominant members of the bacterial community from the water column sample (Figure 3, green columns). The more surprising feature of these data is evident when examining the similarities and differences among the rare members of the community. Although similarity among sediment samples did decrease as the number of sequences per tag decreased, even among those tags present fewer than five times in over 20 000 sequences per sample, there was a remarkable degree of similarity (∼44%). If variability within the community composition of the rare sediment microbes was large then the chance sequencing of identical rare tags would be low, resulting in low similarity among replicate samples. By contrast, the similarity among sediment samples (∼44%) is so much greater than the similarity between the sediment and the water column out-group (2.1±0.3%) that it cannot be explained by the chance sequencing of equally rare taxa. Rather, one possible explanation to explain this degree of similarity is that bacteria in the sediments appear to be under some functional selection that promotes cohesion even among the rare members of the community. Biases associated with the use of different methods for DNA extraction between the sediment samples and the out-group water column sample could compound the differences between the out-group water column and the sediment samples. The essential point here, however, is not that the sediments and water column are different from one another; this is to be expected. Rather, it is the degree of similarity within the rare biosphere of the sediment samples, which demonstrates that repeatable patterns in community composition can be determined and can, in theory, be used as a baseline from which to infer changes in microbial communities across environmental gradients.
When including all the taxonomic data, an unweighted Pair Group Mean Analysis (UPGMA) phenogram shows one cluster of sediment samples that are only 20–30% dissimilar from one another but that is >80% dissimilar to the out-group water column sample (Figure 4a). As a further test of whether the rare biosphere of similar samples could be distinguished from the rare biosphere of an out-group sample, we also performed the UPGMA on taxa present fewer than five times (Figure 4b). The UPGMA clusters of the rare taxa show a slightly different order of clustering than when all sequences were considered (Figure 4a), but nonetheless all sediment samples cluster together and are far removed from the out-group. This provides further evidence that environmental replicates display similar community compositions, even among the rare members of the consortia.
Microbial communities that have fundamentally different structures would not only cluster differently from one another, they would likely have different cumulative frequency distributions. Although it is possible that two samples could have different community compositions but similar frequency distributions, the inverse is not, that is, communities that have different cumulative frequency distributions cannot have the same community structure. Quantifying the shape of the frequency distribution can thus provide a mechanism for confirming differences in community compositions that may result from environmental perturbation. We characterized the frequency distribution of the sediment samples by fitting Gaussian curves to the data (Figure 5). The amplitude, mean and s.d.’s of these curves can then be used to compare among replicates and to contrast with the out-group sample. The sediment replicates had similarly shaped curves and overlapping 95% confidence intervals (CI) (Table 3). Averaged across all the sediment samples, the amplitude of the Gaussian curves indicates that the sediment samples had ∼4000 OTUs (4168±314) compared with 1056 OTUs in the out-group, thus confirming our previous conclusion that these sediment samples harbor considerably greater diversity than was found in the water column draining an adjacent marsh.
The mean and s.d. of the Gaussian curve fits, indicators of the number of sequences per tag and the spread of the data, respectively, were higher in the water column out-group than in the sediment samples (Table 3, Figure 5). This would be expected from a sample that is dominated by a handful of very abundant taxa. The sediment samples, however, contain fewer very high abundance tags; rather, they have a more even distribution of less abundant taxa. This is evident by the different extent of the curves along the x axis (Figure 5). In the sediments, it takes 250–300 of the most abundant tags to account for 50% of all the sequences; in the water column, just the two most dominant tags account for 50% of all the sequences.
Both the sediment samples and the water column out-group sample demonstrate a long tail of low abundance taxa, but this tail is considerably longer in the sediment samples. This is indicated both by the overall taxonomic richness (Table 1) as well as by the Gaussian curve fits. The location of the y intercept on each of the curves indicates the number of sequences that occur only one time (Figure 5). This particular water column sample had 625 tags that occurred once, compared with between 1750 and 2250 tags in the sediment samples. Furthermore, the initial slope of the curves suggest that there are many more tags in the sediments that are present between 2 and 10 times as compared with the water column sample. This analysis underscores both the vast richness of the microbial reservoir in marine sediments and the similar composition of the communities among environmental replicates.
Conclusions
The development of pyrosequencing as a technique for deep sequencing of microbial communities has contributed a tremendous amount of new information to our knowledge of the diversity of these systems. Microbial ecologists are now able to use this technology to begin asking questions about the role that diversity has in understanding ecosystem function. However, the interpretability of these data depends on the magnitude of the variability within environmental replicates, and the degree to which incomplete sequencing exacerbates this variability. The data presented here indicate that despite incomplete sequencing, at least in these salt marsh sediments, the microbial community is surprisingly homogeneous. Individually collected sediment cores had similar estimates of richness and diversity, and similarity indices calculated from sequence information from all the individually collected sediments were of the same magnitude. Furthermore, homogenizing multiple sediment samples in an effort to decrease the variability among individual samples proved unnecessary. The highly similar community structure of the environmental replicates stands in contrast to the wide divergence seen between the sediment samples and an out-group sample collected from a nearby water column. The pyrosequencing method was able to easily differentiate this out-group from the sediment samples.
References
Baas Becking LGM . (1934) Geobiologie of Inleiding tot de Milieukunde. WP Van Stockum & Zoon: The Hague, The Netherlands.
Bowen JL, Ward BB, Morrison HG, Hobbie JE, Valiela I, Deegan LA et al. (2011). Microbial community composition in sediments resists perturbation by nutrient enrichment. ISME J 5: 1540–1548.
Brazelton WJ, Ludwig KA, Sogin ML, Andreishcheva EN, Kelley DS, Shen C-C et al. (2010). Archaea and bacteria with surprising microdiversity show shifts in dominance over 1000-year time scales in hydrothermal chimneys. Proc Natl Acad Sci USA 107: 1612–1617.
Bunge J, Woodard L, Böhning D, Foster JA, Connolly S, Allen HK . (2012). Estimating population diversity with CatchAll. Bioinformatics 28: 1045–1047.
Carson JK, Campbell L, Rooney D, Clipson N, Gleeson DB . (2009). Minerals in soil select distinct bacterial communities in their microhabitats. FEMS Microbiol Ecol 67: 381–388.
Galand PE, Casamayor EO, Kirchman DL, Lovejoy C . (2009). Ecology of the rare microbial biosphere of the Arctic Ocean. Proc Natl Acad Sci USA 106: 22427–22432.
Gihring TM, Green SJ, Schadt CW . (2012). Massively parallel rRNA gene sequencing exacerbates the potential for biased community diversity comparisons due to variable library sizes. Envion Microbiol 14: 285–290.
Hong S-H, Bunge J, Jeon S-O, Epstein SS . (2006). Predicting microbial species richness. Proc Natl Acad Sci USA 103: 117–122.
Howes BL, Howarth RW, Teal JM, Valiela I . (1981). Oxidation-reduction potentials in a salt marsh: spatial patterns and interactions with primary production. Limnol Oceanogr 26: 350–360.
Huber JA, Mark Welch DB, Morrison HG, Huse SM, Neal PR, Sogin ML . (2007). Microbial population structures in the deep marine biosphere. Science 318: 97–100.
Huse SM, Huber JA, Morrison HG, Sogin ML, Mark Welch DB . (2007). Accuracy and quality of massively parallel DNA pyrosequencing. Gen Biol 8: R143.
Huse SM, Dethlefsen L, Huber JA, Mark Welsh DB, Relman DA, Sogin ML . (2008). Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genetics 4: e1000255.
Huse SM, Mark Welch DB, Morrison HG, Sogin ML . (2010). Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol 12: 1889–1898.
Jørgensen BB, Boetius A . (2007). Feast and famine—microbial life in the deep sea bed. Nature Rev Microbiol 5: 770–781.
Kunin V, Engelbrektson A, Ochman H, Hugenholtz P . (2009). Wrinkles in the rare biosphere: pyrosequencing errors lead to artificial inflation of diversity estimates. Environ Microbiol 12: 118–123.
Lehmann J, Solomon D, Kinyangi J, Dathe L, Wirick S, Jacobsen C . (2008). Spatial complexity of soil organic matter forms at nanometer scales. Nature Geosci 1: 238–242.
Ley RE, Harris JK, Wilcox J, Spear JR, Miller SR, Bebout BM et al. (2006). Unexpected diversity and complexity from the Guerrero Negro hypersaline microbial mat. Appl Environ Microbiol 72: 3685–3695.
Lozupone CA, Knight R . (2007). Global patterns in bacterial diversity. Proc Natl Acad Sci USA 104: 11436–11440.
Lozupone CA, Knight R . (2008). Species divergence and the measurement of microbial diversity. FEMS Microbiol Rev 32: 557–578.
Magurran AE . (1988) Ecological Diversity and its Measurement. Princeton University Press: Princeton, NJ.
Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA et al. (2005). Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380.
Morales SE, Cosart TF, Johnson JV, Holben WE . (2009). Extensive phylogenetic analysis of a soil bacterial community illustrates extreme taxon evenness and the effects of amplicon length, degree of coverage, and DNA fractionation on classification and ecological parameters. Appl Environ Microbiol 75: 668–675.
Quince C, Curtis TP, Sloan WT . (2008). The rational exploration of microbial diversity. ISME J 10: 997–1006.
Quince C, Lanzén A, Curtis TP, Davenport RJ, Hall N, Head IM et al. (2009). Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods 6: 639–641.
Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ . (2011). Removing noise from pyrosequenced amplcons. BMC Bioinformatics 12: 38.
Roesch LFW, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, Kent AD et al. (2007). Pyrosequencing enumerates and contrasts soil microbial diversity. ISMEJ 1: 283–290.
Schloss PD . (2008). Evaluating different approaches that test whether microbial communities have the same structure. ISME J 2: 265–275.
Schloss PD, Wescott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. (2009). Introducing MOTHUR: open-source, platform-independent, community supported software for describing complex microbial communities. Appl Environ Microbiol 75: 7537–7541.
Schloss PD, Gevers D, Wescott SL . (2011). Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA based studies. PLoS One 6: e273.
Sogin ML, Morrison HG, Huber JA, Mark Welch DB, Huse SM, Neal PR et al. (2006). Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. Proc Natl Acad Sci USA 103: 12115–12120.
Turnbaugh PJ, Hamaday M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE et al. (2009). A core gut microbiome in obese and lean twins. Nature 457: 480–484.
Valiela I, Teal JM . (1979). The nitrogen budget of a salt marsh. Nature 280: 652–656.
Valiela I, Cole ML . (2002). Comparative evidence that salt marshes and mangroves may protect seagrass meadows from land-derived nitrogen loads. Ecosystems 5: 92–102.
Acknowledgements
Funding for this research came from the National Science Foundation (DEB- 0717155 to JEH and HGM, and award DBI-0400819 to JLB). Support for the sequencing facility comes from the National Institutes of Health and National Science Foundation (NIH/NIEHS 1 P50 ES012742-01 and NSF/OCE 0430724-J Stegeman PI to HGM and MLS) and a grant from the WM Keck Foundation (to MLS). Salary support was contributed by a fellowship from the Princeton University Council on Science and Technology to JLB. Anonymous reviewers improved the quality of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Bowen, J., Morrison, H., Hobbie, J. et al. Salt marsh sediment diversity: a test of the variability of the rare biosphere among environmental replicates. ISME J 6, 2014–2023 (2012). https://doi.org/10.1038/ismej.2012.47
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2012.47
Keywords
This article is cited by
-
Microbial Community Succession Along a Chronosequence in Constructed Salt Marsh Soils
Microbial Ecology (2023)
-
Correlative SIP-FISH-Raman-SEM-NanoSIMS links identity, morphology, biochemistry, and physiology of environmental microbes
ISME Communications (2022)
-
Longitudinal patterns in sediment type and quality during daily flow regimes and following natural hazards in an urban estuary: a Hurricane Harvey retrospective
Environmental Science and Pollution Research (2022)
-
Mechanistic strategies of microbial communities regulating lignocellulose deconstruction in a UK salt marsh
Microbiome (2021)
-
Aggregate sizes regulate the microbial community patterns in sandy soil profile
Soil Ecology Letters (2021)