Abstract
Here, we investigated 124 stinkbug species representing 20 families and 5 superfamilies for their Burkholderia gut symbionts, of which 39 species representing 6 families of the superfamilies Lygaeoidea and Coreoidea were Burkholderia-positive. Diagnostic PCR surveys revealed high frequencies of Burkholderia infection in natural populations of the stinkbugs, and substantial absence of vertical transmission of Burkholderia infection to their eggs. In situ hybridization confirmed localization of the Burkholderia in their midgut crypts. In the lygaeoid and coreoid stinkbugs, development of midgut crypts in their alimentary tract was coincident with the Burkholderia infection, suggesting that the specialized morphological configuration is pivotal for establishment and maintenance of the symbiotic association. The Burkholderia symbionts were easily isolated as pure culture on standard microbiological media, indicating the ability of the gut symbionts to survive outside the host insects. Molecular phylogenetic analysis showed that the gut symbionts of the lygaeoid and coreoid stinkbugs belong to a β-proteobacterial clade together with Burkholderia isolates from soil environments and Burkholderia species that induce plant galls. On the phylogeny, the stinkbug-associated, environmental and gall-forming Burkholderia strains did not form coherent groups, indicating host–symbiont promiscuity among these stinkbugs. Symbiont culturing revealed that slightly different Burkholderia genotypes often coexist in the same insects, which is also suggestive of host–symbiont promiscuity. All these results strongly suggest an ancient but promiscuous host–symbiont relationship between the lygaeoid/coreoid stinkbugs and the Burkholderia gut symbionts. Possible mechanisms as to how the environmentally transmitted promiscuous symbiotic association has been stably maintained in the evolutionary course are discussed.
Similar content being viewed by others
Introduction
Many insects possess symbiotic microorganisms of mutualistic nature either in the gut lumen, in the body cavity or inside the cells. In general, such microbial partners are maintained through host generations by specialized transmission mechanisms (Buchner, 1965; Bourtzis and Miller, 2003; Kikuchi, 2009; Bright and Bulgheresi, 2010). For example, an intracellular symbiont Buchnera in aphids is providing essential amino acids for the host, indispensable for the host growth and reproduction, and transovarially transmitted to eggs or embryos in the maternal host body (Douglas, 1998; Miura et al., 2003). Meanwhile, insects harboring extracellular gut symbionts of mutualistic nature, like termites and stinkbugs, have evolved posthatch mechanisms for symbiont transmission, such as superficial microbial contamination of eggs (egg smearing) and probing of parental microbe-containing excrement (coprophagy) (reviewed in Buchner, 1965). In utero symbiont transmission via milk gland secretion in tsetse flies (Attardo et al., 2008) and capsule-mediated symbiont transmission in plataspid stinkbugs (Hosokawa et al., 2005) represent unique posthatch transmission mechanisms. In these and other diverse insects, the mechanisms for symbiont transmission thus far described are mostly vertical ones from parents to offspring.
On the other hand, environmental symbiont transmission, wherein newborn host organisms acquire their mutualistic partners from environmental microbial pool every generation, has been known from diverse marine invertebrates including luminescent squids (Nyholm and McFall-Ngai, 2004), photosynthetic corals (Baker, 2003), chemoautotrophic tubeworms (Dubilier et al., 2008) and many others. In the aquatic ecosystem, free-living symbionts are protected from environmental stresses such as UV irradiation and desiccation that must be detrimental to free-living symbionts in the terrestrial ecosystem, to which the paucity of environmental symbiont transmission among insects may be relevant. Even in the terrestrial ecosystem, however, some plants are mutualistically associated with specific soil microbes, as in legume–Rhizobium and alder–Frankia nitrogen-fixing symbioses, wherein environmental symbiont transmission occurs (Denarie et al., 1992; Benson and Silvester, 1993). It should be noted that in these cases, the symbionts outside the host organisms are also protected from the above-mentioned environmental stresses in the soil. In this context, it is remarkable as well as understandable that environmental symbiont transmission was recently discovered in an insect–microbe mutualism, wherein the symbiont was soil bacteria of the genus Burkholderia (Kikuchi et al., 2007).
Members of the suborder Heteroptera, known as true bugs or stinkbugs, comprise a large group of insects with incomplete metamorphosis. The infraorder Pentatomomorpha, placed under the Heteroptera, embraces over 12 500 species that are grouped into five superfamilies (Figure 1; Schaefer, 1993; Schuh and Slater, 1995; Henry, 1997). Besides relatively minor predacious and mycophagous species, the majority of pentatomomorphan stinkbugs are plant sap feeders, some of which are notorious as agricultural pests (Schaefer and Panizzi, 2000). In most of the phytophagous species, a number of sac- or tube-like outgrowths (called crypts or caeca) develop in a posterior region of the midgut (so-called midgut fourth section), wherein specific bacteria are harbored (Glasgow, 1914; Miyamoto, 1961; Buchner, 1965; Kikuchi et al., 2008). In the families Pentatomidae, Plataspidae, Acanthosomatidae, Scutelleridae and Parastrachiidae, their gut symbionts belong to distinct lineages in the γ-Proteobacteia, indicating multiple evolutionary origins of the symbiotic associations (Fukatsu and Hosokawa, 2002; Hosokawa et al., 2006, 2010; Prado et al., 2006; Kikuchi et al., 2009; Prado and Almeida, 2009a, 2009b; Kaiwa et al., 2010). In several species of the families Pentatomidae, Plataspidae, Acanthosomatidae, Parastrachiidae and Cydnidae, symbiont-free insects suffered retarded growth and/or nymphal mortality, suggesting mutualistic nature of the symbiotic associations (Müller, 1956; Huber-Schneider, 1957; Schorr, 1957; Abe et al., 1995; Fukatsu and Hosokawa, 2002; Hosokawa et al., 2006; Kashima et al., 2006; Kikuchi et al., 2009; Prado and Almeida, 2009b). All these gut symbionts are vertically transmitted through host generations either via egg smearing in the Pentatomidae, Acanthosomatidae and Scutelleridae (Abe et al., 1995; Prado et al., 2006; Kikuchi et al., 2009; Prado and Almeida, 2009b; Kaiwa et al., 2010), via coprophagy in the Cydnidae (Schorr, 1957), or via capsule transmission in the Plataspidae (Hosokawa et al., 2005).
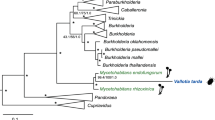
Phylogenetic relationship of the pentatomomorphan stinkbug families. The tree is based on the nuclear 18S rRNA gene phylogeny by Xie et al. (2005) and the mitochondrial whole-genome phylogeny by Hua et al. (2008). Both studies strongly supported the phylogenetic relationship (Aradoidea + (Pentatomoidea + (Pyrrhocoroidea + (Coreoidea + Lygaeoidea)))). Names of stinkbug superfamilies are depicted on the right side. Names of stinkbug families associated with the Burkholderia symbionts are shown in colors. An arrow indicates the hypothetical evolutionary origin of the stinkbug–Burkholderia association.
The bean bug Riptortus pedestris (or Riptortus clavatus as synonym) is known as pest of leguminous crops in eastern Asia (Schaefer and Panizzi, 2000). Whereas the above-mentioned stinkbug families belong to the superfamily Pentatomoidea, R. pedestris is a member of the family Alydidae in the superfamily Coreoidea (see Figure 1). Similar to the pentatomoid stinkbugs, R. pedestris possesses numerous crypts in the posterior midgut, but the bacterium in the symbiotic organ is quite different, belonging to the genus Burkholderia in the β-Proteobacteria (Kikuchi et al., 2005). Aposymbiotic insects suffered marked reduction in body size and weight, indicating mutualistic nature of the symbiosis (Kikuchi et al., 2007). However, distinct from the pentatomoid symbionts thus far studied, the Burkholderia symbiont was present in the rhizosphere of the food plants of R. pedestris, easily culturable on standard microbiological media, and orally acquired by young nymphs of R. pedestris from the environment without vertical transmission (Kikuchi et al., 2007). The discovery of environmental acquisition of beneficial symbiont without vertical transmission in R. pedestris was unprecedented among insects. As the Burkholderia gut symbiont was also detected in a related alydid species Leptocorisa chinensis (Kikuchi et al., 2005), the case of R. pedestris cannot be an orphan exception. It is of interest whether or not such symbiotic associations are also found in other stinkbug species.
In this study, therefore, we surveyed 124 pentatomomorphan stinkbugs representing 20 families and 5 superfamilies for infection with the Burkholderia symbionts, identified 39 Burkholderia-associated stinkbugs from 6 families and 2 superfamilies, and investigated their phylogenetic relationship, localization in vivo, transmission mode and culturability in vitro.
Materials and methods
Insect materials
Samples of Japanese pentatomomorphan species examined in this study are listed in Table 1 and Supplementary Table S1. These insects were collected in 1995–2007 and preserved in acetone at room temperature until use (Fukatsu, 1999).
DNA extraction
Acetone-preserved adult insects were individually dissected in a phosphate-buffered saline (137 mM NaCl, 8.1 mM Na2HPO4, 2.7 mM KCl, 1.5 mMKH2PO4 (pH 7.5)), from which the midgut fourth section with crypts was collected. For the stinkbug species that possess no midgut crypts, the whole area of the midgut was collected. The dissected tissues were homogenized in 200 μl of lysis buffer (10 mM Tris-HCl (pH 8.0), 1 mM EDTA, 0.1 M NaCl, 0.5% SDS, 0.2 mg ml−1 proteinase K) and incubated at 56 °C for 2 h. The lysate was extracted with 200 μl of phenol:chloroform:isoamyl alcohol (25:24:1). DNA was recovered from the purified lysate by ethanol precipitation, and dissolved in 200 μl of TE buffer (10 mM Tris-HCl (pH 8.0), 0.1 mM EDTA).
Diagnostic PCR
A 0.75 kb region of 16S rRNA gene of Burkholderia spp. was amplified by the specific primers Burk16SF (5′-TTTTGGACAATGGGGGCAAC-3′) and Burk16SR (5′-GCTCTTGCGTAGCAACTAAG-3′) under a temperature profile of 95 °C for 10 min followed by 30 cycles of 95 °C for 30 s, 55 °C for 1 min and 72 °C for 1 min (Kikuchi et al., 2005). To check the quality of template DNA samples, a 0.65 kbp region of insect mitochondrial cytochrome oxidase I gene was amplified with the primers LCO1490 and HCO2198 (Folmer et al., 1994).
Cloning, genotyping and sequencing
A 1.5 kbp region of bacterial 16S rRNA gene was amplified by PCR with the universal primers 16SA1 and 16SB1 under a temperature profile of 95 °C for 10 min followed by 30 cycles of 95 °C for 30 s, 55 °C for 1 min and 72 °C for 1 min (Fukatsu and Nikoh, 1998). The PCR products were cloned with a TA-cloning vector pT7Blue (Takara) and Escherichia coli DH5α competent cells (Takara). To check the length of the inserted DNA fragment, white colonies expected to contain inserted plasmid were directly subjected to PCR with the primers Univ19 (5′-GTTTTCCCAGTCACGACGT-3′) and Rev20 (5′-AGCTATGACCATGATTACGC-3′). When a PCR product of expected size (1.5 kbp) was obtained, the product was digested by restriction endonucleases HaeIII and RsaI, and electrophoresed in agarose gels. Three clones or more from each of all restriction genotypes were cultured and subjected to plasmid extraction using QIAprep-Spin Miniprep Kit (QIAGEN, Chuo-ku, Japan). The purified plasmids were eluted with 50 μl of distilled water and subjected to DNA sequencing with DNA Sequencing Kit FS (PerkinElmer, Chiyoda-ku, Japan) and 3130xl Genetic Analyzer (Applied Biosystems, Chuo-ku, Japan).
Molecular phylogenetic analysis
Multiple alignments of the nucleotide sequences were generated by the program Clustal W (Thompson et al., 1994) and then manually realigned using the program SeaView (Galtier et al., 1996). Neighbor-joining (NJ) trees were constructed using the Clustal W under the Kimura's two-parameter model (Kimura, 1980). Maximum likelihood (ML) trees were estimated by the program Phylo_Win (Galtier et al., 1996) under the HKY substitution model (Hasegawa et al., 1985). Bootstrap tests were performed with 1000 and 100 replications in the NJ and ML analyses, respectively.
In situ hybridization
Two oligonucleotide probes Alsym16S (5′-ACACTCAAAGCCTGCCAGT-3′; Kikuchi et al., 2005) and BURK129 (5′-CCACTACAGGACACGTTC-3′), whose 5′ end was labeled with AlexaFluor555, were used for in situ hybridization. These probes specifically targeted different regions in 16S rRNA of the Burkholderia symbionts, and were simultaneously used for enhancement of the hybridization signals. The tissue samples were incubated in a hybridization buffer (20 mM Tris-HCl (pH 8.0), 0.9 M NaCl, 0.01% SDS, 30% formamide) containing 50 nM each of the probes and 0.5 μM of SYTOX Green (Invitrogen, Chuo-ku, Japan). After an overnight incubation, the samples were thoroughly washed in phosphate-buffered saline and mounted in Slowfade antifade solution (Molecular Probes, Chuo-ku, Japan), and observed under an epifluorescent microscope (Axiophot, Carl Zeiss, Shinjuku-ku, Japan) and a laser scanning confocal microscope (LSCM Pascal5, Carl Zeiss). To confirm specificity of the detection, a series of control experiments were conducted as described (Kikuchi et al., 2005).
Symbiont culture
Field-collected insects were surface-sterilized in 70% ethanol, and individually dissected in phosphate-buffered saline. A midgut region with crypts was carefully cut out, repeatedly washed with fresh sterilized water to minimize possible microbial contamination, and homogenized in 1 ml of fresh sterilized water. An adequate amount of the homogenate was spread on yeast extract and glucose (YG) agar plates (0.5% yeast extract, 0.4% glucose, 0.1% NaCl, 1.5% Bacto agar) and incubated at 26 °C for 3 days or longer. From each of the plates, 15 or more colonies were randomly picked and subjected to PCR, cloning and sequencing of 16S rRNA gene.
Results
General observation of midgut crypts in diverse stinkbugs
We dissected 124 pentatomomorphan stinkbug species and inspected their midgut under a binocular microscope. Development of crypts in the midgut fourth section was observed in 96 species representing 15 families (Table 1). Arrangements of the midgut crypts were distinct among the stinkbug taxa, which were classified into at least four morphological types (Table 1; also see Kikuchi et al., 2008): tubular outgrowths in the Blissidae, Berytidae and most species of the Rhyparochromidae (Figure 2b, c, e, f, h and i); small crypts arranged in two rows in the Coreidae, Alydidae, Plataspidae, Parastrachiidae, Cydnidae, Pachygronthidae and several species of the Rhyparochromidae (Figure 2k, l, n, o, q and r; also see Kikuchi et al., 2005; Hosokawa et al., 2010); small crypts arranged in four rows in the Pentatomidae, Scutelleridae, Urostylidae and Dinidoridae (see Abe et al., 1995; Kaiwa et al., 2010); and a pair of large flat assemblage of crypts in the Acanthosomatidae (see Kikuchi et al., 2009). These results were in agreement with early histological observations of stinkbug alimentary tracts (Glasgow, 1914; Miyamoto, 1961; Buchner, 1965).
Midgut organization in stinkbugs of the superfamilies Lygaeoidea and Coreoidea. (a–c) Yemma exilis (Lygaeoidea: Berytidae); (d–f) Dimorphopterus pallipes (Lygaeoidea: Blissidae); (g–i) Togo hemipterus (Lygaeoidea: Rhyparochromidae); (j–l) Pachygrontha antennata (Lygaeoidea: Pachygronthidae); (m–o) Cletus punctiger (Coreoidea: Coreidae); (p–r) Leptocorisa chinensis (Coreoidea: Alydidae). Adult female insects, dissected midguts and enlarged images of the midgut crypts are shown from left to right. Bars show 1 mm. Abbreviations: M1, midgut first section; M2, midgut second section; M3, midgut third section; M4, midgut fourth section with crypts; M4B, bulbous section before M4; H, hindgut.
Detection of Burkholderia symbionts from diverse stinkbugs
These stinkbug species were subjected to diagnostic PCR using Burkholderia-specific primers. Of the 124 species, Burkholderia infection was detected from 39 species of the families Berytidae, Blissidae, Rhoparochromidae, Pachygronthidae, Coreidae and Alydidae, which belong to the superfamilies Lygaeoidea and Coreoidea. In these stinkbug groups, notably, Burkholderia infection was coincident with development of the midgut crypts: all the 39 species with the midgut crypts were Burkholderia-positive (Table 1; Figure 2), whereas the other 22 species exhibited neither the crypts nor the Burkholderia infection (Table 1; Supplememtary Figure S1). The infection frequencies in natural populations were generally high: 91% (10/11) in Yemma exilis; 100% (31/31) in Dimorphopterus pallipes; 100% (36/36) in Togo hemipterus; 100% (18/18) in Pachygrontha antennata; 100% (3/3) in Acanthocoris sordidus, Hygia lativentris and Homoeocerus unipunctatus and 88% (22/25) in Cletus punctiger (Table 1).
Bacterial 16S rRNA gene sequences from Burkholderia-positive stinkbugs
Of the 39 Burkholderia-positive stinkbugs, selected 19 species were subjected to PCR and cloning of bacterial 16S rRNA gene from their midgut crypts. Restriction fragment length polymorphism profiles of the observed 190 clones, 10 clones each from the 19 samples, exhibited identical patterns (data not shown). Three or more clones from each of the samples were subjected to DNA sequencing. All sequences obtained from a single insect were identical, while the sequences from different species slightly (<0.03%) differed from each other. BLAST searches against the DNA databases revealed that the sequences exhibited the highest (>99%) similarities to 16S rRNA gene sequences of Burkholderia symbionts from alydid stinkbugs R. pedestris and L. chinensis (Kikuchi et al., 2005, 2007).
In vivo localization of Burkholderia symbionts
The following representatives of crypt-bearing lygaeoid/coreoid stinkbugs, D. pallipes (Blissidae), T. hemipterus (Rhyparochromidae), A. sordidus (Coreidae) and L. chinensis (Alydidae), were dissected and subjected to in situ hybridization targeting 16S rRNA of Burkholderia symbionts. In all the species, Burkholderia signals were consistently localized in the crypts associated with the midgut fourth section (Figure 3). The localization patterns were similar to that described for R. pedestris (Alydidae; Kikuchi et al., 2005).
Fluorescent in situ hybridization targeting 16S rRNA of Burkholderia symbionts in dissected midguts of lygaeoid and coreoid stinkbugs. Red signals indicate Burkholderia symbionts, whereas green signals show host insect nuclei. (a) Acanthocoris sordidus (Coreoidea: Coreidae); (b) Leptocorisa chinensis (Coreoidea: Alydidae); (c) Dimorphopterus pallipes (Lygaeoidea: Blissidae); (d) Togo hemipterus (Lygaeoidea: Rhyparochromidae). Bars show 0.5 mm. Abbreviations: M1, midgut first section; M2, midgut second section; M3, midgut third section; M4, midgut fourth section with crypts; M4B, bulbous section before M4; H, hindgut.
Isolation and culture of Burkholderia symbionts
Considering that the Burkholderia symbiont of R. pedestris is culturable on standard bacterial media (Kikuchi et al., 2007), we attempted to isolate Burkholderia symbionts using YG medium agar plates from the following lygaeoid/coreoid stinkbugs: Y. exilis (Berytidae); D. pallipes (Blissidae); P. exiguus and T. hemipterus (Rhyparochromidae); P. antennata (Pachygronthidae); A. sordidus, H. lativentris and C. puntiger (Coreidae); and additionally R. pedestris and L. chinensis (Alydidae). Within 3 days after plating of homogenized midgut crypts, a number of white, circular and convex colonies appeared for all the stinkbug samples examined (Figure 4a–f). Microscopic observation of bacterial cells from the colonies consistently revealed 1.0–2.0 μm rods (Figure 4g–l). When 15 or more of the white colonies were picked from each plate and subjected to diagnostic PCR, all the colonies were diagnosed as Burkholderia-positive. In an individual of P. antennata, several yellowish and brownish colonies were exceptionally observed, whose 16S rRNA gene sequences showed high (>99%) sequence similarities to those of Stenotrophomonas maltophilia (AM743169) and Delftia tsuruhatensis (EF421404), respectively.
Colony forms (a–f) and bacterial cell images (g–l) of Burkholderia symbiont strains YEX123 from Yemma exilis, DPA58 from Dimorphopterus pallipes, THE68 from Togo hemipterus, PAN137 from Pachygrontha antennata, CPU52 from Cletus punctiger and LCH90 from Leptocorisa chinensis. Colony formations on YG agar plates were observed after incubation at 26 °C for 3 days. Stationary-phase bacterial cells were observed by phase-contrast microscopy. Bars show 1 mm in (a–f) and 10 μm in (g–l).
Detection of co-infecting Burkholderia symbionts
The cultured Burkholderia colonies were mostly uniform in morphology. However, it was occasionally found that some colonies obtained from a single insect differed in size and shape. In such cases, two to three colonies were picked for each of the colony types and subjected to PCR and sequencing of 16S rRNA gene. All obtained sequences showed high similarities (97–100%) to 16S rRNA gene sequences of the Burkholderia symbionts identified from dissected midgut crypts of the lygaeoid/coreoid stinkbugs as described above. In total, 24 different Burkholderia strains were isolated from 16 insects representing 10 species and 6 families, among which 10, 4 and 2 insects were identified as single-, double- and triple-infected, respectively (Table 2).
Molecular phylogenetic analysis of Burkholderia symbionts
The 16S rRNA gene sequences of the Burkholderia symbionts were subjected to molecular phylogenetic analysis together with Burkholderia and allied β-proteobacterial 16S rRNA gene sequences retrieved from the DNA databases. The sequences from the lygaeoid and coreoid stinkbugs formed a group in the genus Burkholderia, although the whole assemblage received poor statistical supports (45% for NJ and <40% for ML). In the group, the sequences from the coreid stinkbugs formed a compact and well-supported monophyletic group (100% for both NJ and ML), named the ‘Coreidae clade’. The other sequences were placed in a poorly supported clade (75% for NJ and <40% for ML). The clade also contained the sequences of Burkholderia symbionts from the alydid stinkbugs R. pedestris and L. chinensis (Kikuchi et al., 2005, 2007), environmental Burkholderia isolates such as strains AK-5, NF100 and others (Hayatsu et al., 2000; Takenaka et al., 2003), and gall-forming Burkholderia species such as Candidatus B. calva, Candidatus B. kirkii, Candidatus B. nigropunctata and Candidatus B. verschuerenii from African Psychotria plants (Van Oevelen et al., 2002, 2004). These stinkbug-associated, gall-forming and environmental Burkholderia strains were phylogenetically not coherent, but scattered on the phylogeny in an intermingled manner (Figure 5; Supplementary Figure S2).
Phylogenetic relationship of 43 Burkholderia symbiont genotypes identified from 21 stinkbug species of the superfamilies Lygaeoidea and Coreoidea on the basis of 16S rRNA gene sequences. A neighbor-joining tree, inferred from 1228 unambiguously aligned nucleotide sites, is shown. The Burkholderia symbionts were detected from the stinkbug families Berytidae (orange), Blissidae (brown) Rhyparochromidae (blue) and Pachygronthidae (purple) of the superfamily Lygaeoidea, and the stinkbug families Coreidae (red) and Alydidae (green) of the superfamily Coreoidea. Stars indicate cultured strains of the Burkholderia symbionts, whose strain identifiers are shown in parentheses (also see Table 2). The sequences from Riptortus pedestris and Leptocorisa chinensis reported in a previous study (Kikuchi et al., 2005) are underlined. The bootstrap values >40% are depicted at the nodes. In brackets are shown nucleotide sequence accession numbers. Abbreviation: GS, gut symbiont.
Detection of Burkholderia symbionts from eggs
Eggs of several lygaeoid and coreoid stinkbugs were subjected to diagnostic PCR detection of the Burkholderia symbionts. No Burkholderia signal was detected from all eggs of D. pallipes (Blissidae), P. exiguus (Rhyparochromidae), C. punctiger and H. lativentris (Coreidae), and R. pedestris (Alydidae). Only a small fraction of eggs from T. hemipterus (Rhyparochromidae) and L. chinensis (Alydidae) exhibited a faint signal of Burkholderia (Table 3). None of the stinkbug eggs accompanied symbiont capsule-like particles (data not shown).
Discussion
In previous studies, gut symbiotic bacteria of the genus Burkholderia have been identified from two alydid stinkbug species, R. pedestris and L. chinensis (Kikuchi et al., 2005, 2007). Here, we performed an extensive survey of 124 stinkbug species representing 20 families and 5 superfamilies, and identified Burkholderia symbionts from 39 species representing 6 families and 2 superfamilies, namely Lygaeoidea and Coreoidea (Table 1). Irrespective of the stinkbug groups, the Burkholderia symbionts were localized in the specialized symbiotic organ, namely a posterior midgut section bearing crypts (Figure 3), and infection frequencies in natural populations were generally high (Table 1). These results indicate that the β-proteobacterial Burkholderia symbionts are widely associated with lygaeoid and coreoid stinkbugs.
In the superfamily Pentatomoidea, meanwhile, different groups of stinkbugs are associated with different lineages of γ-proteobacterial midgut symbionts: Ishikawaella with the family Plataspidae (Hosokawa et al., 2006), Rosenkranzia with the family Acanthosomatidae (Kikuchi et al., 2009), and several γ-proteobacterial lineages with the families Pentatomidae (Prado and Almeida, 2009a), Scutelleridae (Kaiwa et al., 2010) and Parastrachiidae (Hosokawa et al., 2010). In the molecular phylogeny of the pentatomomorphan families, the Lygaeoidea/Coreoidea associated with β-proteobacterial Burkholderia symbionts formed a distinct major group from the Pentatomoidea associated with γ-proteobactertial symbionts (Figure 1; Xie et al., 2005; Hua et al., 2008). These patterns suggest that host–symbiont specificity/compatibility may exist at the higher taxomic levels of the stinkbugs, which has been maintained over evolutionary time.
We demonstrated that in the lygaeoid and coreoid stinkbugs, the universal occurrence of Burkholderia does not entail host–symbiont co-speciation. Molecular phylogenetic analyses revealed that the symbiont phylogenies did not reflect the host systematics at all: the same host species frequently harbored different symbiont genotypes, and different host species often possessed nearly or completely identical symbiont genotypes (Figure 5; Supplementary Figure S2). These patterns strongly suggest that host–symbiont associations are generally promiscuous in the Lygaeoidea and the Coreoidea. The situation looks different from the situation in the Pentatomoidea, wherein host–symbiont associations are, if not strict, more or less stable. In the families Plataspidae and Acanthosomatidae, respectively, the symbiont phylogenies are highly congruent with the host phylogenies, indicating stable vertical transmission and co-speciation of their gut symbionts over evolutionary time (Hosokawa et al., 2006; Kikuchi et al., 2009). In the family Pentatomidae, although their gut symbionts are presumably of multiple evolutionary origins, the symbionts are generally conserved within the same species and sometimes between congenic species (Prado and Almeida, 2009a).
Previous works showed that, in different groups of the Pentatomoidea, γ-proteobactertial gut symbionts are vertically transmitted through host generations (Schorr, 1957; Abe et al., 1995; Hosokawa et al., 2005; Prado et al., 2006; Kikuchi et al., 2009; Prado and Almeida, 2009b; Kaiwa et al., 2010). In this study, inspection of eggs of lygaeoid and coreoid stinkbugs revealed no stable vertical transmission of the Burkholderia symbionts to their eggs (Table 3). The host–symbiont phylogenetic promiscuity (Figure 5; Supplementary Figure S2) taken together, we suggest that in the lygaeoid and coreoid stinkbugs, their Burkholderia gut symbionts are generally acquired from the environment every generation. Such host–symbiont phylogenetic promiscuity is commonly found in the symbiotic associations without vertical transmission, such as coral-dinoflagellate, legume-Rhizobium, and legume-Burkholderia symbioses (Young and Haukka, 1996; Baker, 2003; Bontemps et al., 2010). It should be noted that occasional vertical transmission of the symbiont via, for example, accidental fecal contamination of eggs (see Table 3; Huber-Schneider, 1957) is compatible with the environmental acquisition of the symbiont in the same species. Needless to say, the presumable prevalence of the environmental symbiont acquisition should be experimentally verified in each of the lygaeoid and coreoid species, as shown in R. pedestris (Kikuchi et al., 2007).
The global stability and the local promiscuity of the host–symbiont associations in the pentatomomorphan stinkbugs are of evolutionary interest. The host–symbiont promiscuity in the lygaeoid and coreoid stinkbugs must be due to the environmental symbiont acquisition. The polyphyly of the vertically-transmitted symbionts in the pentatomoid stinkbugs are probably caused by occasional horizontal transfers of the symbionts across distinct host lineages. Here it is puzzling why the lygaeoid and coreoid stinkbugs remain associated with the β-proteobacterial symbionts despite the intensive host–symbiont mixing at lower levels, so do the pentatomoid stinkbugs with the γ-proteobacterial symbionts. Although speculative, the host–symbiont affinities mediated by their surface molecules might be relevant to the mechanisms for specific recognition and establishment of the symbiosis at the midgut crypts.
Biological roles of the Burkholderia symbionts for the lygaeoid and coreoid stinkbugs have been poorly understood. The only experimental proof was obtained for the alydid species R. pedestris, wherein aposymbiosis resulted in reduced body size (Kikuchi et al., 2007). On account of the specific localization in highly developed symbiotic organ (Figure 2) and the universal occurrence and conservation among the diverse lygaeoid and coreoid species (Table 1), it appears likely that the Burkholderia symbionts generally have important roles for their insect hosts. Considering that all the Burkholderia-associated stinkbugs were phytophagous (Table 1), their symbionts might provide nutritional components such as vitamins and essential amino acids that are often deficient in the plant sap diet. Some Burkholderia strains exhibit nitrogen-fixing ability (Estrada-De et al., 2001), and the Burkholderia symbionts might contribute nitrogenous metabolites to their stinkbug hosts. On account of the taxonomic breadth and diversity of the lygaeoid and coreoid stinkbugs, the effects of the Burkholderia symbionts might be different from species to species, possibly ranging from commensalistic through beneficial to essential for their insect hosts. In this context, the Burkholderia symbionts of the ‘Coreidae clade’ (Figure 5; Supplementary Figure S2) are of particular interest, because the distinct symbiont lineage is specifically associated with coreid species, representing an exception of the generally promiscuous host–symbiont associations in the lygaeoid and coreoid stinkbugs. We suggest the possibility that an advanced form of stinkbug-Burkholderia mutualism would be identified in these coreid species.
In this study, we found that not all the lygaeoid and coreoid species harbor the Burkholderia gut symbionts. In the Lygaeoidea, stinkbugs of the Berytidae, Blissidae, Rhyparochromidae and Pachygronthidae were associated with the Burkholderia symbionts, whereas species of the Malcidae, Lygaeidae and Geocoridae were not. In the Coreoidea, stinkbugs of the Coreidae and Alydidae possessed the Burkholderia symbionts, whereas species of the Rhopalidae did not (Table 1). It is currently elusive what ecological, physiological and/or phylogenetic factors are relevant to the presence/absence of the Burkholderia symbionts. It should be noted that, in all the lygaeoid and coreoid stinkbugs, the presence of the Burkholderia symbiont was coincident with the presence of crypts in the posterior midgut (Figure 2; Supplementary Figure S1; Table 1), suggesting that the specialized morphological configuration is pivotal for establishment and maintenance of the symbiotic association. As the presence/absence of the crypts/Burkholderia reflected the host insect systematics (Table 1), it appears plausible that the symbiotic trait is mainly determined by the host genotype. On the other hand, the possibility cannot be ruled out that the Burkholderia infection is also involved in the formation of the symbiotic structure in the host midgut. In leguminous plants, infection with symbiotic Rhizobium induces nodule formation in the root (Perret et al., 2000; Oldroyd and Downie, 2008). In Euprymna squids, infection with luminescent Vibrio fischeri is required for normal morphogenesis of the symbiotic light organ (McFall-Ngai and Ruby, 1991; Nyholm and McFall-Ngai, 2004). Whether or not the Burkholderia symbiont is involved in the formation of the host symbiotic organ is of interest, deserving future experimental and developmental studies on symbiotic and aposymbiotic insects.
The soil environment harbors an enormous diversity of microorganisms (Garbeva et al., 2004), wherein the fraction of Burkholderia symbionts must be very small. Hence, it is expected that the nymphs of lygaeoid and coreoid stinkbugs orally acquire diverse microorganisms, from which the Burkholderia symbiont is selected to establish specific infection in the midgut crypts. In symbiotic associations entailing environmental transmission in general, the host must have some mechanisms for enrichment and harvesting of specific microorganisms. Once the symbiosis is established, there must be mechanisms to ensure that the association is stable so that the symbionts do not overgrow the host and the host does not eliminate the symbiont (Nyholm and McFall-Ngai, 2004). In the squid–Vibrio and legume–Rhizobium symbioses, intricate cellular and molecular crosstalks have been shown to underlie the establishment and maintenance of the symbiotic associations (Perret et al., 2000; Koropatnick et al., 2004; Nyholm and McFall-Ngai, 2004; Oldroyd and Downie, 2008). In the stinkbug–Burkholderia symbiosis, biochemical and transcriptomic analyses of the midgut crypts would provide insights into what molecular and cellular processes are operating at the host–symbiont interface.
PCR, cloning and sequencing of 16S rRNA gene from a dissected midgut identified a single sequence type, indicating that a single symbiont genotype tends to dominate the gut microbiota in each of the Burkholderia-infected lygaeoid and coreoid stinkbugs. However, culturability of the Burkholderia symbiont enabled identification of hidden diversity of the symbiont genotypes coexisting in the same insects: infrequent colonies of different morphotypes on agar plates often represented different Burkholderia genotypes (Table 2). Such multiple infections seem to be a natural outcome of the environmental symbiont acquisition, which are also found in other systems such as legume–Rhizobium, coral–dinoflagellate and tubeworm–chemoautotroph mutualisms (Moawad and Schmidt, 1987; Rowan et al., 1997; Baker, 2003; Dubilier et al., 2008).
The consequences of hosting multiple symbionts have been a recurrent theme in the evolutionary biology of mutualism. Such multiple infections create a potential tragedy of the commons (Hardin, 1968), which can destabilize and collapse mutualism because within-host competitions tend to favor less beneficial, cheater genotypes (Bull, 1994; West et al., 2002). Hence, it is of interest why mutualisms can be maintained under the absence of vertical transmission and the frequent mixing of different symbiont genotypes. A possible mechanism is so-called ‘partner choice’, wherein the host recognizes benevolent symbiont genotypes and selectively establishes association with them. Another possible mechanism is so-called ‘sanctions’, wherein the host monitors the performance of different symbiont genotypes and punishes less beneficial ones (Maynard Smith and Szathmáry, 1995; Wilkinson and Sherratt, 2001; Kiers and Denison, 2008). In the soybean–Rhizobium association, an experimental support for host sanctions against less beneficial symbionts has been provided (Kiers et al., 2003). On the soybean root, a number of nodules are formed, each of which may represent a symbiont genotype. Such structured distribution of different symbiont genotypes enables effective operation of host sanctions. In this context, we point out that numerous crypts in the posterior midgut of stinkbugs are regarded as similar structural compartments. It is of interest whether different Burkholderia genotypes are separately harbored in different crypts, and whether the crypts harboring different symbiont genotypes tend to exhibit different levels of growth and development.
Overall, the findings presented in this study illuminate dazzlingly promiscuous host–symbiont associations between the diverse lygaeoid/coreoid stinkbugs and the diverse Burkholderia genotypes. It is hypothesized that multiple host species may interact with multiple symbiont genotypes, potentially exchanging the partners every generation as follows: different Burkholderia genotypes are present in the soil environment, are acquired by nymphal lygaeoid/coreoid stinkbugs, proliferate in the midgut crypts, are released from the insects via excrement or cadaver, spend a free-living life in the soil, and are acquired by nymphal stinkbugs of the same or different species, and so forth. Of course, there must be some permissive range in the host–symbiont associations. Each of the stinkbug species may either be more prone to infection, or perform better, with specific Burkholderia genotypes, and vice versa. In the enormous space of possible host–symbiont combinations, the host–symbiont permissive ranges may be stratified, overlapped and nested in a complicated manner. As the environmental symbiont acquisition requires host–symbiont encounters, the symbiotic associations must be affected by the habitat and the ecology of the respective symbiotic partners, which are influenced by environmental factors such as geography, climate and vegetation. Here, despite the many-to-many rather than one-to-one relationships, the lygaeoid/coreoid stinkbugs and the Burkholderia bacteria have established a form of mutualism, which has been maintained, if not so strictly, during the evolutionary course of the insect superfamilies. Such ecological and evolutionary perspectives are strikingly different from those conventionally inferred from the well-studied insect–microbe endosymbioses represented by aphid–Buchnera and tsetse–Wigglesworthia relationships, which broadens our understanding of possible evolutionary trajectories leading to stable mutualistic associations.
References
Abe Y, Mishiro K, Takanashi M . (1995). Symbiont of brown-winged green bug, Plautia stali Scott. Jpn J Appl Entomol Zool 39: 109–115.
Attardo GM, Lohs C, Heddi A, Alam UH, Yildirim S, Aksoy S . (2008). Analysis of milk gland structure and function in Glossina morsitans: milk protein production, symbiont populations and fecundity. J Insect Physiol 54: 1236–1242.
Baker AC . (2003). Flexibility and specificity in coral-algal symbiosis: diversity, ecology, and biogeography of Symbiodinium. Annu Rev Ecol Evol Syst 34: 661–689.
Benson DR, Silvester WB . (1993). Biology of Frankia strains, actinomycete symbionts of actinorhizal plants. Microbiol Rev 57: 293–319.
Bontemps C, Elliott GN, Simon MF, Dos Reis Junior FB, Gross E, Lawton RC et al. (2010). Burkholderia species are ancient symbionts of legumes. Mol Ecol 19: 44–52.
Bourtzis K, Miller TA . (2003). Insect symbiosis. CRC press: Boca Raton, FL.
Bright M, Bulgheresi S . (2010). A complex journey: transmission of microbial symbionts. Nat Rev Microbiol 8: 218–230.
Buchner P . (1965). Endosymbiosis of animals with plant microorganisms. Interscience: New York, USA.
Bull JJ . (1994). Virulence. Evolution 48: 1423–1437.
Denarie J, Debelle F, Rosenberg C . (1992). Signaling and host range variation in nodulation. Annu Rev Microbiol 46: 497–531.
Douglas AE . (1998). Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu Rev Entomol 43: 17–37.
Dubilier N, Bergin C, Lott C . (2008). Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat Rev Microbiol 6: 725–740.
Estrada-De P, Bustillos-Cristales R, Caballero-Mellado J . (2001). Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. Appl Environ Microbiol 67: 2790–2798.
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R . (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Marine Biol Biotechnol 3: 294–299.
Fukatsu T . (1999). Acetone preservation: a practical technique for molecular analysis. Mol Ecol 8: 1935–1945.
Fukatsu T, Nikoh N . (1998). Two intracellular symbiotic bacteria from the mulberry psyllid Anomoneura mori (Insecta, Homoptera). Appl Environ Microbiol 64: 3599–3606.
Fukatsu T, Hosokawa T . (2002). Capsule-transmitted gut symbiotic bacterium of the Japanese common plataspid stinkbug, Megacopta punctatissima. Appl Environ Microbiol 68: 389–396.
Galtier N, Gouy M, Gautier C . (1996). SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Bioinformatics 12: 543–548.
Garbeva P, van Veen JA, van Elsas JD . (2004). Microbial diversity in soil: selection microbial populations by plant and soil type and implications for disease suppressiveness. Annu Rev Phytopathol 42: 243–270.
Glasgow H . (1914). The gastric caeca and the caecal bacteria of the Heteroptera. Biol Bull 3: 101–171.
Hardin G . (1968). The tragedy of the commons. Science 162: 1243–1248.
Hasegawa M, Kishino H, Yano T . (1985). Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22: 160–174.
Hayatsu M, Hirano M, Tokuda S . (2000). Involvement of two plasmids in fenitrothion degradation by Burkholderia sp. strain NF100. Appl Environ Microbiol 66: 1737–1740.
Henry TJ . (1997). Phylogenetic analysis of family groups within the infraorder pentatomomorpha (Hemiptera: Heteroptera), with emphasis on the Lygaeoidea. Ann Entomol Soc Am 90: 275–301.
Hosokawa T, Kikuchi Y, Meng XY, Fukatsu T . (2005). The making of symbiont capsule in the plataspid stinkbug Megacopta punctatissima. FEMS Microbiol Ecol 54: 471–477.
Hosokawa T, Kikuchi Y, Nikoh N, Meng XY, Hironaka M, Fukatsu T . (2010). Phylogenetic position and peculiar genetic traits of the midgut bacterial symbiont in the stinkbug Parastrachia japonensis. Appl Environ Microbiol 76: 4130–4135.
Hosokawa T, Kikuchi Y, Nikoh N, Shimada M, Fukatsu T . (2006). Strict host-symbiont cospeciation and reductive genome evolution in insect gut bacteria. PLoS Biol 4: e337.
Hua J, Li M, Dong P, Cui Y, Xie Q, Bu W . (2008). Comparative and phylogenomic studies on the mitochondrial genomes of Pentatomomorpha (Insecta: Hemiptera: Heteroptera). BMC Genomics 9: 610.
Huber-Schneider L . (1957). Morphologische und physiologische Untersuchungen an der Wanze Mesocerus marginatus L. und ihren Symbionten (Heteroptera). Z Morph Ökol Tiere 46: 433–480.
Kaiwa N, Hosokawa T, Kikuchi Y, Nikoh N, Meng XY, Kimura N et al. (2010). Primary gut symbiont and secondary Sodalis-allied symbiont in the scutellerid stinkbug Cantao ocellatus. Appl Environ Microbiol 76: 3486–3494.
Kashima T, Nakamura T, Tojo S . (2006). Uric acid recycling in the shield bug, Parastrachia japonensis (Hemiptera: Parastrachiidae), during diapause. J Insect Physiol 52: 816–825.
Kiers ET, Denison RF . (2008). Sanctions, cooperation, and the stability of plant-rhizosphere mutualisms. Annu Rev Ecol Evol Syst 39: 215–236.
Kiers ET, Rousseau RA, West SA, Denison RF . (2003). Host sanctions and the legume-rhizobium mutualism. Nature 425: 78–81.
Kikuchi Y . (2009). Endosymbiotic bacteria in insects: their diversity and culturability. Microbes Environ 24: 195–204.
Kikuchi Y, Meng XY, Fukatsu T . (2005). Gut symbiotic bacteria of the genus Burkholderia in the broad-headed bugs Riptortus clavatus and Leptocorisa chinensis (Heteroptera: Alydidae). Appl Environ Microbiol 71: 4035–4043.
Kikuchi Y, Hosokawa T, Fukatsu T . (2007). Insect-microbe mutualism without vertical transmission: a stinkbug acquires beneficial gut symbiont from environment every generation. Appl Environ Microbiol 73: 4308–4316.
Kikuchi Y, Hosokawa T, Fukatsu T . (2008). Diversity of bacterial symbiosis in stinkbugs. Microbial Ecology Research Trends Dijk, TV (ed). Nova Science Publishers, Inc: New York, USA, pp. 39–63.
Kikuchi Y, Hosokawa T, Nikoh N, Meng XY, Kamagata Y, Fukatsu T . (2009). Host-symbiont co-speciation and reductive genome evolution in gut symbiotic bacteria of acanthosomatid stinkbugs. BMC Biol 7: 2.
Kimura M . (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120.
Koropatnick TA, Engle JT, Apicella MA, Stabb EV, Goldman WE, McFall-Ngai MJ . (2004). Microbial factor-mediated development in a host-bacterial mutualism. Science 306: 1186–1188.
Maynard Smith J, Szathmáry E . (1995). The Major Transitions in Evolution. Oxford: Freeman.
McFall-Ngai MJ, Ruby EG . (1991). Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science 254: 1491–1494.
Miura T, Braendle C, Shingleton A, Sisk G, Kambhampati S, Stern DL . (2003). A comparison of parthenogenetic and sexual embryogenesis of the pea aphid Acyrthosiphon pisum (Hemiptera: Aphidoidea). J Exp Zool B 295: 59–81.
Miyamoto S . (1961). Comparative morphology of alimentary organs of Heteroptera, with the phylogenetic consideration. Sieboldia 2: 197–259.
Moawad M, Schmidt EL . (1987). Occurrence and nature of mixed infections in nodules of field-grown soybeans (Glycine max). Biol Fertil Soils 5: 112–114.
Müller HJ . (1956). Experimentelle Studien an der Symbiose von Coptosoma scutellatum Geoffr (Hem. Heteropt.). Z Morph Ökol Tiere 44: 459–482.
Nyholm SV, McFall-Ngai MJ . (2004). The winnowing: establishing the squid-Vibrio symbiosis. Nat Rev Microbiol 2: 632–642.
Oldroyd GE, Downie JA . (2008). Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59: 519–546.
Perret X, Staehelin C, Broughton WJ . (2000). Molecular basis of symbiotic promiscuity. Microbiol Mol Biol Rev 64: 180–201.
Prado SS, Almeida RP . (2009a). Phylogenetic placement of pentatomid stink bug gut symbionts. Curr Microbiol 58: 64–69.
Prado SS, Almeida RP . (2009b). Role of symbiotic gut bacteria in the development of Acrosternum hilare and Murgantia histrionica. Entomol Exp Appl 132: 21–29.
Prado SS, Rubinoff D, Almeida RPP . (2006). Vertical transmission of a pentatomid caeca-associated symbiont. Ann Entomol Soc Am 99: 577–585.
Rowan R, Knowlton N, Baker A, Jara J . (1997). Landscape ecology of algal symbionts creates variation in episodes of coral bleaching. Nature 388: 265–269.
Schaefer CW . (1993). The Pentatomomorpha (Hemiptera: Heteroptera): an annotated outline of its systematic history. Eur J Entomol 90: 105–122.
Schaefer CW, Panizzi AR . (2000). Heteroptera of Economic Importance. CRC Press: Florida, USA.
Schorr H . (1957). Zur Verhaltensbiologie und Symbiose von Brachypelta aterrima Först (Cydnidae, Heteroptera). Z Morph Ökol Tiere 45: 561–602.
Schuh RT, Slater JA . (1995). True bugs of the world (Hemiptera: Heteroptera). Cornell University Press: New York, USA.
Takenaka S, Okugawa S, Kadowaki M, Murakami S, Aoki K . (2003). The metabolic pathway of 4-aminophenol in Burkholderia sp. strain AK-5 differs from that of aniline and aniline with C-4 substituents. Appl Environ Microbiol 69: 5410–5413.
Thompson JD, Higgins DG, Gibson TJ . (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680.
Van Oevelen S, De Wachter R, Vandamme E, Robbrecht E, Prinsen E . (2002). Identification of the bacterial endosymbionts in leaf galls of Psychotria (Rubiaceae, angiosperms) and proposal of ‘Candidatus Burkholderia kirkii’ sp. nov. Int J Syst Evol Microbiol 52: 2023–2027.
Van Oevelen S, De Wachter R, Vandamme E, Vandamme P, Robbrecht E, Prinsen E . (2004). ‘Candidatus Burkholderia calva and ‘Candidatus Burkholderia nigropunctata’ as leaf gall endosymbionts of African Psychotria. Int J Syst Evol Microbiol 54: 2237–2239.
West SA, Kiers ET, Pen I, Denison RF . (2002). Sanctions and mutualism stability: when should less beneficial mutualists be tolerated? J Evol Biol 15: 830–837.
Wilkinson DM, Sherratt TN . (2001). Horizontally acquired mutualisms, an unsolved problem in ecology? Oikos 92: 377–384.
Xie Q, Bu W, Zheng L . (2005). The Bayesian phylogenetic analysis of the 18S rRNA sequences from the main lineages of Trichophora (Insecta: Heteroptera: Pentatomomorpha). Mol Phylogenet Evol 34: 448–451.
Young JPW, Haukka KE . (1996). Diversity and phylogeny of rhizobia. New Phytol 133: 87–94.
Acknowledgements
We thank H Higuchi, M Hironaka, N Ijichi, M Ishizaki, K Ito, T Kashima, K Kohno, S Kudo, F Kuchiki, M Takai, T Takemoto and T Yasuda for providing insect samples. This study was funded by the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN). YK and TH were supported by the Research Fellowship of the Japan Society for the Promotion of Science (JSPS) for Young Scientists.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Kikuchi, Y., Hosokawa, T. & Fukatsu, T. An ancient but promiscuous host–symbiont association between Burkholderia gut symbionts and their heteropteran hosts. ISME J 5, 446–460 (2011). https://doi.org/10.1038/ismej.2010.150
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2010.150
Keywords
This article is cited by
-
Soil microbiomes and one health
Nature Reviews Microbiology (2023)
-
Species diversity of environmentally-transmitted bacteria colonizing Riptortus pedestris (Hemiptera: Alydidae) and symbiotic effects of the most dominant bacteria
Scientific Reports (2023)
-
Symbioses shape feeding niches and diversification across insects
Nature Ecology & Evolution (2023)
-
Obligate Gut Symbiotic Association with Caballeronia in the Mulberry Seed Bug Paradieuches dissimilis (Lygaeoidea: Rhyparochromidae)
Microbial Ecology (2023)
-
Host-Adapted Strains of Spodoptera frugiperda Hold and Share a Core Microbial Community Across the Western Hemisphere
Microbial Ecology (2023)