Abstract
The relative abundance of transcripts encoding proteins involved in inorganic carbon concentrating mechanisms (CCM), detoxification of reactive oxygen species (ROS) and photosynthesis in the thermophilic cyanobacterium Synechococcus OS-B′ was measured in hot spring microbial mats over two diel cycles, and was coupled with in situ determinations of incoming irradiance and microenvironmental dynamics of O2 and pH. Fluctuations in pH and O2 in the mats were largely driven by the diel cycle of solar irradiance, with a pH variation from ∼7.0 to ∼9.5, and O2 levels ranging from anoxia to supersaturation during night and day, respectively. Levels of various transcripts from mat cyanobacteria revealed several patterns that correlated with incident irradiance, O2 and pH within the mat matrix. Transcript abundances for most genes increased during the morning dark–light transition. Some transcripts remained at a near constant level throughout the light period, whereas others showed an additional increase in abundance as the mat underwent transition from low-to-high light (potentially reflecting changes in O2 concentration and pH), followed by either a decreased abundance in the early afternoon, or a gradual decline during the early afternoon and into the evening. One specific transcipt, psbA1, was the lowest during mid-day under high irradiance and increased when the light levels declined. We discuss these complex in situ transcriptional patterns with respect to environmental and endogenous cues that might impact and regulate transcription over the diel cycle.
Similar content being viewed by others
Introduction
In light-exposed extreme environments, such as hypersaline ponds or geothermal springs, perennial surface-associated microbial communities form densely populated mm–cm thick mats composed of an exopolymer matrix harboring a strictly microbial assemblage of photosynthetic primary producers and heterotrophic microorganisms (Teske and Stahl, 2002). Owing to the absence of bioturbation and grazing by metazoans, such mats are ideal model systems for ecophysiological and ecological studies of microbes, their metabolic interactions, adaptation and evolution (Ward et al., 2008).
Mass transfer in cyanobacterial mats is strongly diffusion limited, causing development of steep chemical gradients (de Beer and Kühl, 2001; Wieland et al., 2001), and strong light attenuation typically confines the euphotic zone from sub mm to a few mm thick layer in most mats (Kühl et al., 1997). As a consequence of a high density of oxygenic phototrophs and the impedance of mass transfer within the exopolymer matrix, there is O2 supersaturation, high pH and marked CO2 depletion in the upper layers of photosynthetic microbial mats under high irradiance (de Beer et al., 1997; Wieland and Kühl, 2006). These conditions may result in photorespiration, severe CO2 limitation, as well as significant photoinhibition and reactive oxygen species (ROS) formation. However, microbial mats can sustain high rates of photosynthesis under high irradiance, suggesting that the cyanobacteria in the mats have efficient inorganic carbon concentrating mechanisms (CCM) and can either control the production of ROS or ameliorate its potential toxic effects. Yet, both the presence and regulation of CCM activity and ROS detoxification are virtually unstudied in mat-dwelling cyanobacteria (cf. recent reviews by Price et al., 2008; Latifi et al., 2009; but see Kilian et al. (2007)).
Studies of CCM in cyanobacteria, its regulation by CO2 levels, the ways in which it has been tailored to growth in specific environments and how it impacts other cellular reactions, including those that generate ROS, have mostly been performed with the freshwater planktonic model organisms Synechocystis sp. PCC 6803 and Synechococcus sp. PCC 7942 (Price et al., 2008; Raven et al., 2008). These organisms have been maintained in culture for decades under laboratory conditions in which there is little fluctuation in pH, O2 and CO2 levels.
Studies of the ways in which cyanobacteria adjust to low CO2 conditions have primarily focused on the CCM, which allows cells to concentrate inorganic carbon (Ci; in this study defined as CO2 and HCO3−) against a concentration gradient and to generate a high concentration of CO2 around ribulose-1,5-bisphosphate carboxylase (Tchernov et al., 2003; Kaplan et al., 2008). Cyanobacteria possess a number of different genes that are induced during Ci deprivation, which allow cells to acquire and fix Ci more efficiently (Volokita et al., 1984; Badger and Price, 1990; Omata et al., 1999; Shibata et al., 2001; Maeda et al., 2002; Price et al., 2004; Zhang et al., 2004; Koropatkin et al., 2007; Folea et al., 2008; Xu et al., 2008); these low Ci-induced genes encode polypeptides that are considered as components of the CCM (Kaplan et al., 2008). The majority of ribulose-1,5-bisphosphate carboxylase in cyanobacteria is localized to a specialized microcompartment, the carboxysome, and the structural and catalytic components of the carboxysome are also considered as part of the CCM (Price et al., 2008).
The high O2 tension found within cyanobacterial mats during mid-day may result in elevated ROS generation (Jamieson et al., 1986). Levels of ROS may also increase as a consequence of both ultraviolet irradiation and excess excitation of the photosynthetic apparatus by high levels of photosynthetically active radiation (400–700 nm). ROS can severely damage cellular components, including the photosynthetic apparatus itself (Aro et al., 2005; Bouchard et al., 2006). Photoinhibition, a consequence of photodamage to photosystem II, can lead to a further rise of ROS levels. Oxidative stress in phototrophs can be alleviated by the synthesis of various enzymes (for example, glutathione peroxidase, glutathione S-transferases, thioredoxins and superoxide dismutase) that help minimize ROS-elicited damage (Goyer et al., 2002; Sarkar et al., 2005; Dayer et al., 2008). Harmful effects of light on phototrophs will also depend on Ci availability, as well as on environmental levels of other nutrients, for example, nitrogen, phosphorus, sulfur and iron (Takahashi and Murata, 2008).
The microbial mats in the effluent channels of the alkaline siliceous hot springs, Mushroom and Octopus Springs (Yellowstone National Park, WY, USA), have been the subject of detailed biogeochemical and microbiological investigations for >40 years (Brock, 1967; Ward et al., 1998, 2006). At temperatures >55°C, these mats are exclusively colonized by prokaryotes, including oxygenic photoautotrophic cyanobacteria in the genus Synechococcus spp. and anoxygenic filamentous phototrophic bacteria, Roseiflexus spp. and Chloroflexus spp.; many other functional groups inhabit these microbial ecosystems, but the photosynthetic microbes are dominant inhabitants (Ward et al., 1998, 2006; Bhaya et al., 2007). Two ecologically relevant cyanobacteria, Synechococcus OS-B′ and Synechococcus OS-A (designated Syn OS-B′ and Syn OS-A), have been isolated from the mats of Mushroom and Octopus Springs (Allewalt et al., 2006), and their genomes were sequenced along with metagenomic sequencing of DNA isolated from microbes in the top layer of the microbial mat (Bhaya et al., 2007). This genomic information provides a strong foundation for studies of cyanobacterial in situ gene expression over the diel cycle (Steunou et al., 2006, 2008).
We investigated the in situ dynamics of the abundance of transcripts from Syn OS-B′ (and cyanobacteria with similar genes) encoding proteins involved in CCM, photosynthesis and ROS detoxification, to resolve diel expression patterns and gain insights into mechanisms controlling expression of these genes in mat-dwelling, thermophilic cyanobacteria. These studies were combined with detailed mapping of temperature, incident irradiance and microenvironmental analysis of pH and O2 conditions in the mat over the diel cycle.
Materials and methods
Sample site and collection
Sampling and in situ measurements were performed over two diel cycles on 30 June–1 July 2005 and 13–14 September 2008 in an effluent channel of Mushroom Spring, Yellowstone National Park. Temperature at the sampling site was 56–61°C during each diel cycle. The downwelling quantum irradiance was measured continuously with a datalogger equipped with a quantum irradiance sensor (LI-1400D, LI190, LI-COR Biosciences, Lincoln, NE, USA). Mat samples (∼1.0 cm2) were collected, the top 2 mm were excised, cut in half and frozen in liquid N2, as described previously (Steunou et al., 2008). Although extensive sampling in 2005 was allocated toward the dark-to-light and light-to-dark transition periods, more samples were collected during daytime in 2008.
Microsensor measurements
Microprofiles of the O2 concentration in the microbial mat were measured in situ in vertical steps of 50–200 μm with a Clark-type O2 microelectrode (Revsbech, 1989) connected to a battery-operated pA meter (PA2000, Unisense A/S, Århus, Denmark), as described in detail elsewhere (Steunou et al., 2006, 2008). Based on data from numerous O2 microprofiles, isopleth diagrams of O2 levels in the mat over the diel cycle were generated with the software package Origin Pro 7.5 (OriginLab Corp., Northampton, MA, USA).
Microprofiles of pH were measured in situ using a pH glass microelectrode (Thomas, 1978; Kühl and Revsbech, 2001) and a standard calomel reference electrode connected to a high impedance mV-meter (VF-4, WPI Inc., Sarasota, FL, USA). Linear calibrations of the mV readings were performed in standard buffers at pH 4.0, 7.0 and 10.0 at in situ temperatures, yielding near-Nernstian sensor responses of 54-57 mV per pH unit change.
Gene annotation
The genome of Syn OS-B′ was examined to identify genes encoding proteins involved in CCM and detoxification of ROS; resources used to identify these genes were Genbank annotation, KEGG genomic data (Kanehisa and Goto, 2000) and homologies to well studied genes in model organisms. Primers used to analyze transcript abundances (Table 1) were designed to specifically target transcripts from Syn OS-B′, based on full genome sequence information (NC_007776 for Syn OS-B′ and NC_007775 for Syn OS-A; Bhaya et al., 2007), and homologous sequences likely associated with variant mat cyanobacteria observed in the metagenomic database (http://blast.jcvi.org/yellowstone-blast/index.cgi?project=gym; Bhaya et al., 2007). Primers were tested on genomic DNA from an axenic culture of Syn OS-B′ and a unicyanobacterial culture of Syn OS-A, as positive and negative controls, respectively. All primers were specific except for those used to quantify transcripts from CYB_1611 (putative bicA), which amplified the same fragment from the genomes of Syn OS-A and Syn OS-B′.
RNA extraction and DNAse treatment
RNA from microbial mat samples collected in 2005 was extracted as described (Steunou et al., 2008). A slightly modified protocol was used to extract the RNA from samples collected in 2008. In this protocol the initial vortexing and cell harvesting steps were removed. Instead, mat pieces were disrupted at full power in the bead-beater for 15–30 s after the addition of 250 μl NaOAc and lysis buffer (see steps 1, 3 in Supplementary Methods 1). We also extracted the sample once with 1 vol chloroform after the final phenol:chloroform extraction, as described in Supplementary Methods 1.
Reverse transcription–quantitative PCR
Random primers (Invitrogen Corp., Carlsbad, CA, USA) were annealed to 1 μg of total RNA and extended for 10 min (25 °C) and 30 min (55 °C) using Transcriptor Reverse Transcriptase (Roche Diagnostics Corp., Indianapolis, IN, USA), according to the manufacturer's instructions. The synthesized complementary DNA was diluted 10-fold and 2 μl of the diluted complementary DNA served as template for quantitative PCR using the FastStart SYBR Green Master system (Roche Diagnostics Corp.) and the Engine Opticon System (Bio-Rad Laboratories Inc., Hercules, CA, USA) according to the manufacturer's instructions. Specific amplification of target genes was 1 cycle at 95 °C for 10 min, and 40 cycles at 94 °C for 10 s, 60 °C or 64 °C for 15 s, 72 °C for 30 s, 80 °C for 10 s, and a final incubation for 10 min at 72 °C during the last cycle. The relative abundance of individual transcripts was determined by using the same amount of RNA in the reverse transcription-reactions and by normalizing each time point over the diel cycle to the lowest amplification signal for each specific transcript (Whelan et al., 2003; Steunou et al., 2008).
Results and discussion
Gene annotation
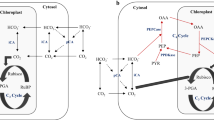
Investigated genes of Syn OS-B′ and the gene clusters in which they occur are shown in Figure 1. Amino acid identities to putative orthologs encoded by the various reference cyanobacterial genomes (Synechococcus sp. PCC 7942; Synechococcus sp. PCC 7002 and Synechocystis sp. PCC 6803) are shown in Supplementary Table 1. In all cases, the deduced protein sequences were most identical to homologs in Syn OS-A, except for CrtO, which is not present on the Syn OS-A genome. Additional information regarding genes selected for expression analysis can be found in Supplementary Information 1 and Supplementary Figures 1 and 2.
Clusters of genes analyzed for in situ gene expression over two diel cycles in Mushroom Spring (bold type). Genes involved in CCM and Ci fixation (dotted or green arrows), detoxification of reactive oxygen species (ROS; black or red arrows), putative regulators (horizontal lines or yellow arrows) and photosynthetic electron transport (oblique or blue arrows) in Synechococcus OS-B′ (genome accession number NC_007776). Gray arrows indicate genes not known to be directly or indirectly involved in these functions, and white arrows encode hypothetical proteins. The color reproduction of this figure is available on the html full text version of the manuscript.
Environmental analysis
The O2 and pH levels in the uppermost mat layers (∼50–150 μm depth) closely tracked the incident irradiance, where O2 fluctuated between anoxia during the night, suboxic conditions during a prolonged period of dim light in the early morning, O2 supersaturation during most of the day, and a period of fluctuating oxic and suboxic conditions during late afternoon (Figure 2). The pH in the mat spanned >2 units, from pH ∼7.0 during the night (2005 and 2008) to pH ∼9.2 (2008) and ∼9.3–9.8 (2005) during periods of high light in the day; the pH of the overlying water changed much less (Figure 2, bottom panel). The pronounced daytime increase in pH indicates severe CO2 depletion in the photic zone during periods of high photosynthetic CO2 fixation as reported in studies of other microbial mats (de Beer et al., 1997).
In situ measurements of incident downwelling irradiance (μmol photons m−2 s−1; top panels), O2 levels (% air saturation; central panels) and pH (bottom panels) at different mat depths, as measured over two diel cycles at a 60°C sampling site in Mushroom Spring in July 2005 (left panels) and September 2008 (right panels).
In situ gene expression
Our in situ investigations of diel gene expression patterns involved monitoring levels of specific cyanobacterial transcripts associated with CCM, ROS detoxification and photosynthesis in the microbial mats. The abundance of these transcripts changed significantly in concert with the extremely dynamic physico-chemical conditions of the microbial mat. Analyses of these patterns allow insights into regulatory mechanisms that modulate physiological activities and growth of mat-dwelling thermophilic cyanobacteria.
CCM
Transcripts encoding proteins involved in bicarbonate and CO2 uptake (chpY, cmpA, putative bicA and chpX) increased in the mat during the transition from total darkness to low light (Figure 3), and stayed at this level throughout the low-light period (irradiance of <200 μmol photons m−2 s−1) in the early morning (Figures 3b-e). A further increase in the levels of the chpY, cmpA, putative bicA and chpX transcripts was observed as irradiance levels rose (08:00 and 10:00 hours in 2005 and 2008, respectively). In the early afternoon, the levels of chpY, cmpA and bicA transcripts declined (14:00 hours in 2005 and 15:00 hours in 2008), even though irradiance, O2 concentration and pH were still elevated (Figures 2 and 3a). The chpX transcript also declined somewhat in the early afternoon, although it tended to be maintained at a higher level relative to the other transcripts (Figure 3e).
Incident downwelling irradiance (400-700 nm) (a; μmol photons m−2 s−1) and relative abundance of transcripts from CCM genes (b; chpY, c; cmpA, d; putative bicA, e; chpX, f; ccmM, ccmN, rbcL and g; putative ccmR), as measured over two diel cycles at a 60 °C sampling site in Mushroom Spring in July 2005 (left panels) and September 2008 (right panels). Transcript abundance data show the mean±s.e. (n=3).
These first observations of diel patterns of transcript abundances from genes encoding cyanobacterial Ci transporters in hot spring mats are largely consistent with what has been observed for cultured cyanobacteria. The darkness to light transition elicited an increase in abundance of CCM transcripts; such results could be explained by photoreceptor control or changes in the redox state of the cells (for example, the plastoquinone pool) upon initial light exposure. Both possibilities are in accord with the finding that active Ci-transport is light/energy dependent (Ogawa et al., 1985; Li and Canvin, 1998; Tchernov et al., 2001), and studies of laboratory cultures of Synechocystis sp. PCC 6803 have demonstrated that light is a prerequisite for elevation of CCM transcript levels (McGinn et al., 2003). The additional increase in CCM transcript levels under the low-to-high irradiance transition has a number of potential explanations:
(1) Several investigations on changes in CCM transcript levels suggest that a limiting internal Ci pool can serve as a signal that activates gene expression during illumination (Omata et al., 1999, 2001; Shibata et al., 2001; McGinn et al., 2003; Woodger et al., 2003, 2005). This possibility is plausible for mat cyanobacteria as, during the shift from low-to-high light, the pH in the upper layers of the microbial mat increased from ∼7.5 to >9.0 (Figure 2), indicating a severe depletion of the Ci pool. Depletion of external Ci as a consequence of high photosynthetic demand for Ci by the cyanobacteria would lead to a marked depletion of internal Ci, at least until the CCM proteins facilitate Ci uptake against the concentration gradient. However, Ci limitation, as well as limitation for other nutrients, can also cause a decline in the utilization of reductants produced by photosynthetic electron transport, which in turn might lead to elevated redox levels, ROS production and cellular damage (Takahashi and Murata, 2008); all of these factors would likely impact gene expression.
(2) Part of the elevated CCM transcript levels may also reflect a further light-induced activation of genes in Syn OS-B′ variants that reside at the bottom of the cyanobacterial layer of the mat; these organisms may not have experienced light saturation of gene activity during the low-light period in the morning owing to strong light attenuation in the microbial mat (Ward et al., 2006).
(3) Increased transcript levels may also reflect elevated energy levels required to fuel gene activity in the cyanobacteria and an increase in the cellular redox state with increasing photosynthetic activity as morning progresses (McGinn et al., 2004); these features are also related to (4) elevation of the O2 concentration in the mat (Figure 2), which would impact cyanobacterial respiration, photosynthesis and energy status (Woodger et al., 2005). It is also possible that specific metabolites like 2-phosphoglycolate, accumulating under increasing mat O2 concentrations, could act as co-inducers of gene expression (Marcus et al., 1983; Nishimura et al., 2008). (5) High pH within the microbial mat during midday (Figure 2) may lead to elevated intracellular pH (Ritchie, 1991, 2008) and a potential shift in the intracellular CO2/HCO3− ratio, which might impact gene activity; this ratio may also be influenced by the intracellular ionic composition and carbonic anhydrase activity.
The pronounced decline in CCM transcript levels in the early afternoon as observed for chpY, cmpA and bicA (Figure 3), despite high irradiance and an elevated O2 concentration and pH (Figure 2), suggests that the system may be able to autoregulate its gene expression as a consequence of induced CCM activity and efficient acquisition of Ci. Changes in levels of other cellular metabolites and circadian signals could also have regulatory consequences. The decreasing abundance of transcripts for the Ci transporters during the night may reflect both a marked decrease in light levels and an increase in both internal and external Ci levels as photosynthetic CO2 fixation stops.
Interestingly, the chpX transcript, reported to be constitutively expressed in cyanobacterial cultures (McGinn et al., 2003; Woodger et al., 2003), accumulated in the mat during early morning as well as under the low-to-high light shift, and remained elevated in the afternoon unlike other CCM genes, which declined in the early afternoon (Figure 3). Both irradiance and altered Ci availability may impact chpX transcript differently relative to the other CCM genes, but it is also possible that chpX mRNA degradation is slower than that of other transporter mRNAs. Culture studies of Syn OS-B′ gene expression might help distinguish such differences, although differences in specific transcript abundances observed in situ in the mat and in cell cultures are not unexpected, owing to our limited ability to establish laboratory conditions that mimic the biotic and abiotic conditions in microbial mats.
Genes encoding carboxysome assembly proteins (cmmM, ccmN) and ribulose-1,5-bisphosphate carboxylase (rbcL) increased somewhat pre-dawn (reaching the highest night value at ∼04:00 and ∼05:30 hours in 2005 and 2008, respectively), as well as after the low-to-high light transition (Figure 3f, after 10:00) for both samplings. Carboxysomes are evenly segregated between dividing cells (Savage et al., 2010), and a pre-dawn increase in transcript abundance may prepare the cells for division, which would require division of the carboxysome or for the onset of photosynthetic activity at sunrise. Thus, a circadian element may influence the expression of genes critical for the fixation of CO2 during the day. Such regulation was previously suggested for the rbcL gene based on in situ data (Wyman, 1999), and the levels of ccm and rbcL transcripts (and other transcripts) exhibit circadian oscillations in S. elongatus PCC 7942 (Vijayan et al., 2009).
The rise in ccmM, ccmN and rbcL transcript levels during the low-to-high light transition (Figure 3f) could be explained by the same factors as those discussed above for transcripts encoding the Ci transporters, that is, depletion of internal Ci and an altered redox state, but expression may also be linked to cell division, the timing of which is still unresolved for the hot spring mat cyanobacteria (Ward et al., 1998, 2006). The carboxysome content increases in cultured cyanobacteria after a shift from high-to-low external Ci concentrations (Turpin et al., 1984; McKay et al., 1993), and ccm transcripts have also been shown to increase somewhat in response to low Ci (Woodger et al., 2003). Other studies report no change in the levels of ccm transcripts in response to low Ci-conditions (McGinn et al., 2003), but if the activities of the ccm and rbcL genes are also under circadian control or linked to cell division, the lack of responsiveness could be a consequence of asynchronous cell growth under continuous light.
CcmR, a putative regulator of CCM
Transcripts encoding the putative regulator CcmR markedly increased with the onset of light, and the transcript levels remained relatively high over the entire light period (Figure 3g). The rapid accumulation of transcripts after the onset of light and the near constancy of the transcript level throughout the light period (most apparent in the 2005 samples) suggests that expression of this gene may be modulated by small changes in redox-conditions, photoperception by receptors such as phytochrome, or other cellular processes that are saturated throughout the cyanobacterial layer even at low irradiance. Our results are, to some extent, at odds with results obtained in studies of cmpR/ndhR/ccmR in cultured cyanobacteria (McGinn et al., 2003; Woodger et al., 2003, 2007), where transcript abundances were influenced by Ci levels.
ROS
All transcripts for genes encoding enzymes involved in ROS detoxification increased during the transition from darkness to low light. Some transcripts (sodB, crtP and CYB_2254, which encodes a putative Prx) stayed at a fairly constant level during the low-light period of the early morning (Figure 4) and then increased with increasing irradiance (08:00 and 10:00 hours in 2005 and 2008, respectively); this pattern is similar to that observed for genes involved in Ci uptake. Later in the afternoon, that is, at 14:00 hours, under high irradiance and O2 levels, transcript levels started to decline (sodB, CYB_2254 and crtP). Levels of other ROS-related transcripts (crtO and CYB_0556, encoding a putative glutamate-cysteine ligase) exhibited a relatively small increase over the light period (Figure 4e). The influence of high irradiance on sodB, crtP and crtO transcript levels have been investigated in axenic liquid cultures of Syn OS-B′ (Kilian et al., 2007), and our in situ data are in accord with the results from this laboratory study, in which there was an influence of high light on sodB and crtP transcript levels, whereas the abundance of the crtO transcript appeared independent of irradiance (darkness was not investigated).
Incident downwelling irradiance (400–700 nm) (a; μmol photons m−2 s−1) and relative abundance of transcripts associated with ROS detoxification (b, sodB; c, CYB_2254 which encodes a type 2-Cys Prx; d, crtP; e, crtO and CYB_0556, which encodes a putative glutamate–cysteine ligase), as measured over two diel cycles at a 60 °C sampling site in Mushroom Spring in July 2005 (left panels) and September 2008 (right panels). Transcript abundance data show the mean±s.e. (n=3).
However, the changing irradiance might not directly influence levels of specific transcripts as several variables in the mat environment (for example, O2, ROS levels and pH) and within the cytoplasm of cyanobacteria co-vary with irradiance. The increasing abundance of transcripts encoding ROS-associated proteins during the darkness to low light transition (Figure 4) is likely regulated in a similar way as that of CCM genes; such regulatory features were reported for the sodB gene in Synechocystis sp. PCC 6803 (Kim and Suh, 2005). On the other hand, changes in transcript levels observed during the transition from low-to-high light could be influenced by various factors, many of which have already been discussed for the Ci transporters. Some specific regulatory mechanisms that might be associated with this transition in cyanobacteria are a change in cellular redox state (for example, of the plastoquinone pool), an increased flux of electrons through the cytochrome b6f complex, photosystem I-mediated thioredoxin signaling (Zeller and Klug, 2006; Oelze et al., 2008), and the accumulation of various ROS (Li et al., 2004).
It should be noted that light-induced H2O2 accumulation is not only biologically mediated, but can also be abiotically generated by the action of ultraviolet irradiation on dissolved organic carbon (Wilson et al., 2000b). H2O2 formation and the dynamics that control its accumulation are yet to be investigated in photosynthetic microbial mats. However, there is increased formation of H2O2 in waters flowing over the surface of hot spring microbial mats during the day relative to the night (Wilson et al., 2000a). Such elevated H2O2 from photochemical reactions in the upper layers of microbial mats could significantly impact photosynthetic processes. H2O2 has a relatively long lifetime and can diffuse into cells from the surrounding medium; this contrasts with singlet oxygen which has a short lifetime, although it is often considered to be the most damaging ROS formed by cyanobacteria (Triantaphylides et al., 2008; Triantaphylides and Havaux, 2009).
Based on our in situ data, it is difficult to identify specific regulatory mechanisms that modulate transcript accumulation for ROS-associated genes, as several physiological features of the system are changing simultaneously. However, our data show that at least some genes associated with ROS detoxification (for example, sodB, crtO, CYB_0556) are probably not controlled in the same manner like most CCM genes, as the levels of transcripts from these genes, unlike those encoding most CCM polypeptides, do not decline in the mid-afternoon. Expression patterns of ROS-associated genes appear to largely respond to irradiance and subsequent changes in the mat chemistry (for example, supersaturating O2 levels, high pH and Ci depletion), although they may also be impacted by circadian control.
Photosynthesis
Transcripts from the psbA2/3 genes (reverse transcription quantitative PCR analyses did not distinguish transcripts from these genes because of their near identical nucleotide sequences; two nucleotide differences in coding region) were elevated at the onset of light with the highest levels directly correlating with high incident irradiance (∼1500 μmol photons m−2 s−1; Figures 5a and c). In contrast, the psbA1 transcript levels increased in the early evening and generally declined slowly during the night (Figure 5b). In 2005, the lowest psbA1 transcript levels were observed during the high-light mid-day periods, whereas highest levels were observed under low irradiance in the late afternoon to early evening and in the early morning (Figure 5b). Our in situ results are consistent with culture studies of Syn OS-B′, where psbA2/3 transcript levels increased during the transition from low-to-high light, whereas psbA1 transcript levels increased when the cells were shifted from high-to-low light (Kilian et al., 2007); similar results were reported for other cultivated cyanobacteria (Schaefer and Golden, 1989).
Incident downwelling irradiance (400–700 nm) (a; μmol photons m−2 s−1) and relative abundance of transcripts associated with photosynthetic function (b; psbA1 and c; psbA2/3), as measured over two diel cycles at a 60 °C sampling site in Mushroom Spring in July 2005 (left panels) and September 2008 (right panels). Transcript abundance data show the mean±s.e. (n=3).
The high psbA2/3 transcript levels in the microbial mat during mid-day do not necessarily indicate a higher cyanobacterial D1 protein level than in cyanobacteria experiencing low-light conditions (for example, early morning). The D1 protein is susceptible to ROS/high-light triggered damage and is rapidly degraded when cells are absorbing excess excitation energy (Krieger-Liszkay et al., 2008). A rapid turnover coupled with the fact that ROS may inhibit D1 repair processes (Nishiyama et al., 2001) raises the possibility that the actual level of functional D1 may be lower during high-light exposure in spite of very high transcript accumulation. Our O2 microprofiles show a slight decline in O2 levels in the upper mat layers at around mid-day (Figure 2). However, O2 levels in the mat remained at supersaturating levels throughout the day, indicative of sustained photosynthetic activity, which suggests that the level of psbA2/3 transcripts and the protein encoded by these transcripts are sufficient to ensure the regeneration of active photosystem II reaction centers following damage resulting from high light/ROS.
Conclusion
This study presents the first investigation of in situ transcript abundance of genes involved in CCM and ROS detoxification in conjunction with precise measurements of environmental parameters over a diel cycle in a photosynthetic microbial mat. Gene expression patterns were linked to dynamic shifts of the physico-chemical conditions (pH and O2) in the mat, largely driven by the diel cycle of solar irradiance. The results provide insights into complex in situ variation in gene expression over the course of the day, and point toward potential regulatory mechanisms that control the dynamics of transcript accumulation.
Variations in transcript abundances in Syn OS-B′ under in situ conditions were largely consistent with patterns observed for cyanobacteria grown as axenic liquid cultures under controlled laboratory conditions. This provides validation for the idea that important, ecologically-relevant information can be derived from laboratory studies, although the many factors that co-occur in situ should be considered when performing experiments with axenic cultures in the laboratory. In some cases this complexity, for example, potential integration of circadian control with control through environmental signals, may be lost under culture conditions because of the way in which the cells are grown, that is, continuous light versus light–dark cycling and modulation of light levels over the diel cycle, and differences in features of the surrounding environment including interactions with other microorganisms and the presence of an organic matrix, which can highly impact diffusion rates and lead to the formation of steep chemical gradients. Some of these complexities can be dissected with specific manipulations of axenic cultures, although key aspects of in situ environmental features should always be considered in the interpretation of the data.
Accession codes
References
Allewalt JP, Bateson MM, Revsbech NP, Slack K, Ward DM . (2006). Effect of temperature and light on growth of and photosynthesis by Synechococcus isolates typical of those predominating in the Octopus spring microbial mat community of Yellowstone National Park. Appl Environ Microbiol 72: 544–550.
Aro E-M, Suorsa M, Rokka A, Allahverdiyeva Y, Paakkarinen V, Saleem A et al. (2005). Dynamics of photosystem II: a proteomic approach to thylakoid protein complexes. J Exp Bot 56: 347–356.
Badger MR, Price GD . (1990). Carbon oxysulfide is an inhibitor of both CO2 and HCO3− uptake in the cyanobacterium Synechococcus PCC7942. Plant Physiol 94: 35–39.
Bhaya D, Grossman AR, Steunou A-S, Khuri N, Cohan FM, Hamamura N et al. (2007). Population level functional diversity in a microbial community revealed by comparative genomic and metagenomic analyses. ISME J 1: 703–713.
Bouchard JN, Roy S, Campbell DA . (2006). UVB effects on the photosystem II-D1 protein of phytoplankton and natural phytoplankton communities. Photochem Photobiol 82: 936–951.
Brock TD . (1967). Relationship between standing crop and primary productivity along a hot spring thermal gradient. Ecology 48: 566–571.
Dayer R, Fischer BB, Eggen RIL, Lemaire SD . (2008). The peroxiredoxin and glutathione peroxidase families in Chlamydomonas reinhardtii. Genetics 179: 41–57.
de Beer D, Glud A, Epping E, Kühl M . (1997). A fast-responding CO2 microelectrode for profiling sediments, microbial mats, and biofilms. Limnol Oceanogr 42: 1590–1600.
de Beer D, Kühl M . (2001). Interfacial microbial mats and biofilms. In: Boudreau BP, Jørgensen BB (eds). The Benthic Boundary Layer. Transport Processes and Biogeochemistry. Oxford University Press: New York, pp 374–394.
Folea IM, Zhang P, Nowaczyk MM, Ogawa T, Aro E-M, Boekema EJ . (2008). Single particle analysis of thylakoid proteins from Thermosynechococcus elongatus and Synechocystis 6803: localization of the CupA subunit of NDH-1. FEBS Lett 582: 249–254.
Goyer A, Haslekås C, Miginiac-Maslow M, Klein U, Le Marechal P, Jacquot J-P et al. (2002). Isolation and characterization of a thioredoxin-dependent peroxidase from Chlamydomonas reinhardtii. Eur J Biochem 269: 272–282.
Jamieson D, Chance B, Cadenas E, Boveris A . (1986). The relation of free radical production to hyperoxia. Annu Rev Physiol 48: 703–719.
Kanehisa M, Goto S . (2000). KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28: 27–30.
Kaplan A, Hagemann M, Bauwe H, Kahlon S, Ogawa T . (2008). Carbon acquisition by cyanobacteria: mechanisms, comparative genomics, and evolution. In: Herrero A, Flores E (eds). The Cyanobacteria. Molecular biology, Genomics and Evolution. Caister Academic Press: Norfolk, UK, pp 305–334.
Kilian O, Steunou A-S, Fazeli F, Bailey S, Bhaya D, Grossman AR . (2007). Responses of a thermophilic Synechococcus isolate from the microbial mat of octopus spring to light. Appl Environ Microbiol 73: 4268–4278.
Kim J-H, Suh KH . (2005). Light-dependent expression of superoxide dismutase from cyanobacterium Synechocystis sp. strain PCC 6803. Arch Microbiol 183: 218–223.
Koropatkin NM, Koppenaal DW, Pakrasi HB, Smith TJ . (2007). The structure of a cyanobacterial bicarbonate transport protein, CmpA. J Biol Chem 282: 2606–2614.
Krieger-Liszkay A, Fufezan C, Trebst A . (2008). Singlet oxygen production in photosystem II and related protection mechanism. Photosynth Res 98: 551–564.
Kühl M, Lassen C, Revsbech NP . (1997). A simple light meter for measurements of PAR (400 to 700 nm) with fiber-optic microprobes: application for P vs E0 (PAR) measurements in a microbial mat. Aquat Microb Ecol 13: 197–207.
Kühl M, Revsbech NP . (2001). Biogeochemical microsensors for boundary layer studies. In: Boudreau BP, Jørgensen BB (eds). The Benthic Boundary Layer. Transport Processes and Biogeochemistry. Oxford University Press: New York, pp 180–210.
Latifi A, Ruiz M, Zhang C-C . (2009). Oxidative stress in cyanobacteria. FEMS Microbiol Rev 33: 258–278.
Li H, Singh AK, McIntyre LM, Sherman LA . (2004). Differential gene expression in response to hydrogen peroxide and the putative PerR regulon of Synechocystis sp. strain PCC 6803. J Bacteriol 186: 3331–3345.
Li Q, Canvin DT . (1998). Energy sources for HCO3− and CO2 transport in air-grown cells of Synechococcus UTEX 625. Plant Physiol 116: 1125–1132.
Maeda S-i, Badger MR, Price GD . (2002). Novel gene products associated with NdhD3/D4-containing NDH-1 complexes are involved in photosynthetic CO2 hydration in the cyanobacterium, Synechococcus sp. PCC7942. Mol Microbiol 43: 425–435.
Marcus Y, Harel E, Kaplan A . (1983). Adaptation of the cyanobacterium Anabaena variabilis to low CO2 concentration in their environment. Plant Physiol 71: 208–210.
McGinn PJ, Price GD, Badger MR . (2004). High light enhances the expression of low-CO2-inducible transctipts involved in the CO2-concentrating mechanism in Synechocystis sp. PCC6803. Plant Cell Environ 27: 615–626.
McGinn PJ, Price GD, Maleszka R, Badger MR . (2003). Inorganic carbon limitation and light control the expression of transcripts related to the CO2-concentrating mechanism in the cyanobacterium Synechocystis sp. strain PCC6803. Plant Physiol 132: 218–229.
McKay RML, Gibbs SP, Espie GS . (1993). Effect of dissolved inorganic carbon on the expression of carboxysomes, localization of Rubisco and the mode of inorganic carbon transport in cells of the cyanobacterium Synechococcus UTEX 625. Arch Microbiol 159: 21–29.
Nishimura T, Takahashi Y, Yamaguchi O, Suzuki H, Maeda S, Omata T . (2008). Mechanism of low CO2-induced activation of the cmp bicarbonate transporter operon by a LysR family protein in the cyanobacterium Synechococcus elongatus strain PCC 7942. Mol Microbiol 68: 98–109.
Nishiyama Y, Yamamoto H, Allakhverdiev SI, Inaba M, Yokota A, Murata N . (2001). Oxidative stress inhibits the repair of photodamage to the photosynthetic machinery. EMBO J 20: 5587–5594.
Oelze M-L, Kandlbinder A, Dietz K-J . (2008). Redox regulation and overreduction control in the photosynthesizing cell: complexity in redox regulatory networks. Biochim Biophys Acta 1780: 1261–1272.
Ogawa T, Miyano A, Inoue Y . (1985). Photosystem-I-driven inorganic carbon transport in the cyanobacterium, Anacystis nidulans. Biochim Biophys Acta 808: 77–84.
Omata T, Gohta S, Takahashi Y, Harano Y, Maeda S-I . (2001). Involvement of a CbbR homolog in low CO2-induced activation of the bicarbonate transporter operon in cyanobacteria. J Bacteriol 183: 1891–1898.
Omata T, Price GD, Badger MR, Okamura M, Gohta S, Ogawa T . (1999). Identification of an ATP-binding cassette transporter involved in bicarbonate uptake in the cyanobacterium Synechococcus sp. strain PCC 7942. Proc Natl Acad Sci USA 96: 13571–13576.
Price GD, Badger MR, Woodger FJ, Long BM . (2008). Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J Exp Bot 59: 1441–1461.
Price GD, Woodger FJ, Badger MR, Howitt SM, Tucker L . (2004). Identification of a SulP-type bicarbonate transporter in marine cyanobacteria. Proc Natl Acad Sci USA 101: 18228–18233.
Raven JA, Giordano M, Beardall J . (2008). Insights into the evolution of CCMs from comparisons with other resource acquisition and assimilation processes. Physiol Plant 133: 4–14.
Revsbech NP . (1989). An oxygen microsensor with a guard cathode. Limnol Oceanogr 34: 474–478.
Ritchie R . (1991). Membrane-potential and pH control in the cyanobacterium Synechococcus R-2 (Anacystis nidulans) PCC 7942. J Plant Physiol 137: 409–418.
Sarkar N, Lemaire S, Wu-Scharf D, Issakidis-Bourguet E, Cerutti H . (2005). Functional specialization of Chlamydomonas reinhardtii cytosolic thioredoxin h1 in the response to alkylation-induced DNA damage. Eukaryot Cell 4: 262–273.
Savage DF, Afonso B, Chen AH, Silver PA . (2010). Spatially ordered dynamics of the bacterial carbon fixation machinery. Science 327: 1258–1261.
Schaefer MR, Golden SS . (1989). Differential expression of members of a cyanobacterial psbA gene family in response to light. J Bacteriol 171: 3973–3981.
Shibata M, Ohkawa H, Kaneko T, Fukuzawa H, Tabata S, Kaplan A et al. (2001). Distinct constitutive and low-CO2-induced CO2 uptake systems in cyanobacteria: genes involved and their phylogenetic relationship with homologous genes in other organisms. Proc Natl Acad Sci USA 98: 11789–11794.
Steunou A-S, Bhaya D, Bateson MM, Melendrez MC, Ward DM, Brecht E et al. (2006). In situ analysis of nitrogen fixation and metabolic switching in unicellular thermophilic cyanobacteria inhabiting hot spring microbial mats. Proc Natl Acad Sci USA 103: 2398–2403.
Steunou A-S, Jensen SI, Brecht E, Becraft ED, Bateson MM, Kilian O et al. (2008). Regulation of nif gene expression and the energetics of N2 fixation over the diel cycle in a hot spring microbial mat. ISME J 2: 364–378.
Takahashi S, Murata N . (2008). How do environmental stresses accelerate photoinhibition? Trends Plant Sci 13: 178–182.
Tchernov D, Helman Y, Keren N, Luz B, Ohad I, Reinhold L et al. (2001). Passive entry of CO2 and its energy-dependent intracellular conversion to HCO3− in cyanobacteria are driven by a photosystem I-generated ΔμH+. J Biol Chem 276: 23450–23455.
Tchernov D, Silverman J, Luz B, Reinhold L, Kaplan A . (2003). Massive light-dependent cycling of inorganic carbon between oxygenic photosynthetic microorganisms and their surroundings. Photosynth Res 77: 95–103.
Teske AP, Stahl D . (2002). Microbial mats and biofilms: evolution, structure, and function of fixed microbial communities. In: Staley JT, Reysenbach AL (eds). Biodiversity of Microbial Life: Foundations of Earth’ Biosphere. Wiley-Liss: New York, pp 49–100.
Thomas RC . (1978). Ion-sensitive intracellular microelectrodes: how to make and use them. Academic Press: London, New York.
Triantaphylides C, Havaux M . (2009). Singlet oxygen in plants: production, detoxification and signaling. Trends Plant Sci 14: 219–228.
Triantaphylides C, Krischke M, Hoeberichts FA, Ksas B, Gresser G, Havaux M et al. (2008). Singlet oxygen is the major reactive oxygen species involved in photooxidative damage to plants. Plant Physiol 148: 960–968.
Turpin DH, Miller AG, Canvin DT . (1984). Carboxysome content of Synechococcus leopoliensis (cyanophyta) in response to inorganic carbon. J Phycol 20: 249 253.
Vijayan V, Zuzow R, O’Shea EK . (2009). Oscillations in supercoiling drive circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA 106: 22564–22568.
Volokita M, Zenvirth D, Kaplan A, Reinhold L . (1984). Nature of the inorganic carbon species actively taken up by the cyanobacterium Anabaena variabilis. Plant Physiol 76: 599–602.
Ward DM, Bateson MM, Ferris MJ, Kühl M, Wieland A, Koeppel A et al. (2006). Cyanobacterial ecotypes in the microbial mat community of mushroom spring (Yellowstone National Park, Wyoming) as species-like units linking microbial community composition, structure and function. Phil Trans R Soc B 361: 1997–2008.
Ward DM, Cohan FM, Bhaya D, Heidelberg JF, Kühl M, Grossman A . (2008). Genomics, environmental genomics and the issue of microbial species. Heredity 100: 207–219.
Ward DM, Ferris MJ, Nold SC, Bateson MM . (1998). A natural view of microbial biodiversity within hot spring cyanobacterial mat communities. Microbiol Mol Biol Rev 62: 1353–1370.
Whelan JA, Russell NB, Whelan MA . (2003). A method for the absolute quantification of cDNA using real-time PCR. J Immunol Methods 278: 261–269.
Wieland A, de Beer D, Damgaard LR, Kühl M, van Dusschoten D, Van As H . (2001). Fine-scale measurement of diffusivity in a microbial mat with nuclear magnetic resonance imaging. Limnol Oceanogr 46: 248–259.
Wieland A, Kühl M . (2006). Regulation of photosynthesis and oxygen consumption in a hypersaline cyanobacterial mat (Camargue, France) by irradiance, temperature and salinity. FEMS Microbiol Ecol 55: 195–210.
Wilson CL, Hinman NW, Cooper WJ, Brown CF . (2000a). Hydrogen peroxide cycling in surface geothermal waters of Yellowstone National Park. Environ Sci Tech 34: 2655–2662.
Wilson CL, Hinman NW, Sheridan RP . (2000b). Hydrogen peroxide formation and decay in iron-rich geothermal waters: the relative roles of abiotic and biotic mechanisms. Photochem Photobiol 71: 691–699.
Woodger FJ, Badger MR, Price GD . (2003). Inorganic carbon limitation induces transcripts encoding components of the CO2-concentrating mechanism in Synechococcus sp. PCC7942 through a redox-independent pathway. Plant Physiol 133: 2069–2080.
Woodger FJ, Badger MR, Price GD . (2005). Sensing of inorganic carbon limitation in Synechococcus PCC7942 is correlated with the size of the internal inorganic carbon pool and involves oxygen. Plant Physiol 139: 1959–1969.
Woodger FJ, Bryant DA, Price GD . (2007). Transcriptional regulation of the CO2-concentrating mechanism in a euryhaline, coastal marine cyanobacterium, Synechococcus sp. strain PCC 7002: role of NdhR/CcmR. J Bacteriol 189: 3335–3347.
Wyman M . (1999). Diel rhythms in ribulose-1,5-bisphosphate carboxylase/oxygenase and glutamine synthetase gene expression in a natural population of marine picoplanktonic cyanobacteria (Synechococcus spp.). Appl Environ Microbiol 65: 3651–3659.
Xu M, Ogawa T, Pakrasi HB, Mi H . (2008). Identification and localization of the CupB protein involved in constitutive CO2 uptake in the cyanobacterium, Synechocystis sp. strain PCC 6803. Plant Cell Physiol 49: 994–997.
Zeller T, Klug G . (2006). Thioredoxins in bacteria: functions in oxidative stress response and regulation of thioredoxin genes. Naturwissenschaften 93: 259–266.
Zhang P, Battchikova N, Jansen T, Appel J, Ogawa T, Aro E-M . (2004). Expression and functional roles of the two distinct NDH-1 complexes and the carbon acquisition complex NdhD3/NdhF3/CupA/Sll1735 in Synechocystis sp PCC 6803. Plant Cell 16: 3326–3340.
Acknowledgements
We thank the US National Park Service and personnel from the Yellowstone National Park for their permission to conduct this work (permit YELL-2058; MK) and their kind and helpful assistance. We also thank C Klatt, E Becraft and DM Ward for assistance and logistical help during the fieldtrips, F Fazeli for help in the laboratory, and D Gonzalez-Ballester and S Bailey for helpful inputs and discussions. This study was funded by an internationalization PhD grant from the Danish Research Council (MK), with additional support from the National Science Foundation (Frontiers in Integrative Biology Program Grant EF-0328698; ARG, DB, MK), the Danish Natural Science Research Council (MK) and the Carnegie Institution for Science (DB, ARG).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Jensen, S., Steunou, AS., Bhaya, D. et al. In situ dynamics of O2, pH and cyanobacterial transcripts associated with CCM, photosynthesis and detoxification of ROS. ISME J 5, 317–328 (2011). https://doi.org/10.1038/ismej.2010.131
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2010.131
Keywords
This article is cited by
-
Thermophilic Dehalococcoidia with unusual traits shed light on an unexpected past
The ISME Journal (2023)
-
In situ metabolomic- and transcriptomic-profiling of the host-associated cyanobacteria Prochloron and Acaryochloris marina
The ISME Journal (2018)
-
Trade-offs between microbiome diversity and productivity in a stratified microbial mat
The ISME Journal (2017)
-
Effects of temperature on growth, photophysiology, Rubisco gene expression in Prorocentrum donghaiense and Karenia mikimotoi
Ocean Science Journal (2016)
-
Temporal metatranscriptomic patterning in phototrophic Chloroflexi inhabiting a microbial mat in a geothermal spring
The ISME Journal (2013)