Abstract
Sponges form close relationships with bacteria, and a remarkable phylogenetic diversity of yet-uncultured bacteria has been identified from sponges using molecular methods. In this study, we use a comparative metagenomic analysis of the bacterial community in the model sponge Cymbastela concentrica and in the surrounding seawater to identify previously unrecognized genomic signatures and functions for sponge bacteria. We observed a surprisingly large number of transposable insertion elements, a feature also observed in other symbiotic bacteria, as well as a set of predicted mechanisms that may defend the sponge community against the introduction of foreign DNA and hence contribute to its genetic resilience. Moreover, several shared metabolic interactions between bacteria and host include vitamin production, nutrient transport and utilization, and redox sensing and response. Finally, an abundance of protein–protein interactions mediated through ankyrin and tetratricopeptide repeat proteins could represent a mechanism for the sponge to discriminate between food and resident bacteria. These data provide new insight into the evolution of symbiotic diversity, microbial metabolism and host–microbe interactions in sponges.
Similar content being viewed by others
Introduction
Sponges are ancient sessile filter-feeding metazoans that harbour complex bacterial communities. The considerable bacterial biomass associated with sponges, the existence of sub-populations of bacterial cells in different host compartments with proposed symbiotic host–bacteria interactions and the early discovery of the sponge–bacteria ecosystem as a source of novel bioactives and natural product chemistry have driven many efforts to identify and improve our understanding of sponge-associated microorganisms (Taylor et al., 2007b; Vogel, 2008). Moreover, recent phylogenetic studies suggest that marine sponges harbour specific, stable microbial communities that are distinct in composition from those of the surrounding seawater (Taylor et al., 2007b). The existence of such distinct communities is remarkable given that the sponge's bacterial community is constantly exposed to large amounts of filtered, planktonic bacteria (Vogel, 1977). However, limited success in culturing sponge bacteria has left us with only a rudimentary understanding of the functional nature of the interactions between bacteria and their marine sponge hosts (Webster and Blackall, 2008). Given that the sponge–bacteria association has been presented as one of the prime examples of marine symbiosis (Taylor et al., 2007a), a fundamental insight into the properties of sponge-associated bacteria is required to define their ecological role and their relationship with the sponge host (Taylor et al., 2007a), as well as to understand the extent to which the relationship is a symbiosis.
The sponge Cymbastela concentrica is an abundant marine sponge found in shallow waters along the Australian east coast and has been previously studied with respect to its bacterial community composition (Taylor et al., 2004b, 2005). Fingerprinting analysis based on the 16S rRNA gene of temporal and spatial replicate samples for this sponge showed a stable bacterial community profile, which was distinct from that of the surrounding water, and showed specific phylotypes shared with the bacterial communities of other sponges. Hence, Cymbastela concentrica represents a model system for sponge–bacteria interaction. In this study, we report a comparative metagenomic analysis that leads to the discovery of novel, previously unrecognized functional properties of sponge bacteria.
Materials and methods
Sampling, cell separation and DNA extraction of communities
Water samples and sponge specimen were collected from Botany Bay near Bare Island, Sydney, Australia (S 33.59.461; E 151.13.946) on 18 October 2006 between 10.47 am and 1.15 pm (T1). The physico-chemical parameters of the water column at 2 m depth at the time of sampling were pH: 8.05; temperature: 17.1–18.3 °C; salinity: 33.6 ppt; chlorophyll: 0.82–1.44 mg l−1; and dissolved oxygen: 5.95–7.00 mg l−1. Sampling of the sponge was repeated on 7 February 2007 at approximately 11.30 am (T2) at the same site. The two water samples from T1 (see below) were also subjected to nutrient analysis and the values are given in Supplementary Table S3.
Duplicate samples of 100 l of water were pumped from a depth of approximately 2 m and directly sequentially filtered through 20, 3, 0.8 and 0.1 μm filters using the same equipment and procedure as described in Rusch et al. (2007). DNA was extracted from the filters corresponding to the size fraction that is smaller than 0.8 μm and greater than 0.1 μm as described by Rusch et al. (2007).
Replicate sponge individuals (a few metres apart) of approximately 45–50 g wet weight were collected by self-contained underwater breathing apparatus diving from a depth of about 7 m below the intake site for the water sampling. Samples were placed into filtered sterilized seawater using sterile scalpels and forceps and transported on ice to the laboratory at UNSW (approximately 15 min travelling time) for direct processing. Surface barnacles were removed from the sponge and wet weights were recorded. Throughout the subsequent cell-separation procedure, samples were taken for microscopic observations. Bacterial cells are separated from sponge cells and tissue using a series of filtration and centrifugation steps, as applied in other studies (Schirmer et al., 2005; Fieseler et al., 2006), and DNA was extracted from the separated cells. Details of these procedures are given in the Supplementary Information.
Shotgun sequencing, assembly and sequence analysis
Clone libraries were produced and sequenced from environmental DNA samples as described in Rusch et al. (2007). After samples had passed several quality control steps (including base quality, read length distribution and a check for eukaryotic DNA (as described below)), large-scale sequencing was performed on ABI3730XL sequencers on two water replicates from T1 (named BBAY01 and 02) and one sponge sample each from T1 and T2 (named BBAY04 and 15). Shotgun sequences were assembled essentially as described in Rusch et al. (2007) with the following modifications. The read fragments were assembled with Celera Assembler software (Myers et al., 2000), version 3.1–5.1 from the public repository (SourceForge, http://wgs-assembler.sf.net). A 12% error rate was permitted in the unitigger (utgErrorRate) with a 14% error rate allowed in the overlapper (ovlErrorRate), consensus (cnsErrorRate) and scaffolder (cgwErrorRate) module. Seed length (merSizeOvl) was set to 14. Overlap trimming, extended clear ranges and surrogates were turned on. Fragment correction and bubble popping were turned off. Basic statistics for each library are given in Supplementary Table S1.
Eukaryotic contaminations such as DNA derived from mitochondria might not have been efficiently removed from bacterial cells by the size and density fractionation described above. We therefore filtered our data set by searching (blastN) all DNA fragments against the 21 June 2008 version of (National Center for Biotechnology Information) NCBI NT database. On the basis of these search results, sequences were classified into taxonomic groups using the MEGAN algorithm (Huson et al., 2007) with parameters ‘min score’ set at 30% and ‘top score’ set at 10%. Manual evaluation confirmed that this procedure effectively removes scaffolds and singletons derived from mitochondrial DNA. For planktonic samples, the amount of sequencing classified as ‘Eukaryotes’ corresponded to 1% of the total assembled nucleotides, whereas for sponge samples, roughly 3% of the DNA data were flagged as ‘Eukaryotes’. All open reading frames (ORFs) associated with those putative eukaryotic DNA fragments were disregarded from the functional comparison.
Scaffolds and singletons were processed with Metagene (Noguchi et al., 2006) to identify ORFs. The algorithm implemented in Metagene is specifically designed for prokaryotic (i.e., bacterial and archaeal) gene detection from metagenomic data sets (Noguchi et al., 2006). Each ORF was searched by blastP search against the non-redundant Genbank database (NR) and taxonomically classified with MEGAN with parameters ‘min score’ set at 35% and ‘top score’ set at 10%. For functional annotation, each ORF was searched against the clusters of orthologous group (COG) (Tatusov et al., 2003) and TIGRFAM (Haft et al., 2003) databases using hmmer version 2.3.2 (Eddy, 1998) and against the curated section of SwissProt (Boeckmann et al., 2003) using blastP (Altschul et al., 1990), applying a confidence cutoff of 10−20. The functional annotation of each ORF was multiplied by the average coverage of the DNA fragment to estimate the abundance of each functional assignment; a similar strategy was recently used by Tringe et al. (2008). In addition, ORFs were searched with hmmer (cutoff of 10−20) against a set of 31 conserved, single-copy marker proteins (Ciccarelli et al., 2006). The hits were normalized against the length of each marker model and averaged to obtain an estimate of the number of genome equivalents present in the samples. All functional assignments were divided by this genome equivalent number to give an average abundance of each function per genome. Replicate samples of each biological system (sponge and water) were compared using the t-test (two-tailed distribution, equivalent variance) with a significance cutoff of P<0.05. In addition, only differences in the relative abundance greater than a factor of three between biological samples were considered.
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs) were identified using the programme CRISPRs finder and categorized into long and small/non-perfect CRISPRs according to Grissa et al. (2007). CRISPR numbers were normalized to genome equivalents and statistical significance was determined by t-test.
Transposable elements were further characterized by blastP analysis of all predicted proteins against a hand-curated database for proteins in insertion sequences kindly provided by Mike Chandler, Laboratoire de Microbiologie et Genetique Moleculaires, Toulouse Cedex, France (Siguier et al., 2006). Only hits with greater than 50% identity over at least 50% length of the query peptide were considered for the analysis.
Analysis of the medium subunit of the aerobic-type carbon monoxide dehydrogenase was performed on sequences that matched COG1319 with hmmer E-values of less than 10−20. Sequences greater than 100 amino acids in length (plus references and outgroup) were aligned with ClustalX (Thompson et al., 2002) using default parameters (i.e. Gonnet weighting matrix). Sequences with less than 50% coverage of alignment were removed (6 out of 45) and the matrix was recalculated. Trees were built using the neighbour-joining algorithm incorporated in ClustalX with 1000 bootstraps.
Ankyrin repeat (AR) and tetratricopeptide repeat (TPR) proteins for the sponge samples were assigned to putative taxonomic origins using the lowest common ancestor algorithm implemented in MEGAN (Huson et al., 2007). BlastP searches against NCBI's NR database were parsed with MEGAN parameters with min score=30%, top percentage=10% and min support for taxa=1. All TPR proteins were assigned to the domain bacteria or lower taxons within the domain. Assignments for the 121 unique AR proteins are given in Supplementary Table S4. The possibility that the 36 AR proteins assigned to Trichomonas vaginalis are due to a contamination of the sample with an organism closely related to Trichomonas vaginalis was further investigated by performing the MEGAN analysis as above, but using all proteins in the sponge metagenome. A total of 40 proteins from this data set were assigned to Trichomonas vaginalis, which contains the 36 AR proteins plus four more proteins with similarity to the ankyrin model described by COG0666, but with E-values slightly above our cutoff of 10−20. If a contamination had occurred, one would expect other protein families to be included in the Trichomonas vaginalis assignment of the total data set; hence, we conclude that these 36 proteins are similar to Trichomonas vaginalis, but not derived from it (nor from any close relative). The 121 ankyrin repeat and 66 TPR proteins were further classified by hmmer searches (E<10−5) against the Pfam database (Sammut et al., 2008) and only hits against the ANK and SEL1 modes (ANK and SEL1 corresponds to the AR and TPR in Pfam, respectively) were observed. The number of repeats were found to be between 1 and 37, with the majority of the proteins having between 3 to 7 and 2 to 4 repeats for the AR and TPR proteins, respectively (see Supplementary Figure S5). Secretion of repeat protein was predicted with SignalP, using default parameters for Gram-negative bacteria (Bendtsen et al., 2004). The top three non-redundant blast hits in the NR of the secreted AR protein were used to build a sequence alignment with ClustalX using default parameters (i.e., Gonnet weighting matrices). Trees were built using the neighbour-joining algorithm incorporated in ClustalX with 1000 bootstraps.
Binning
Binning of scaffolds with more than 20 Kb sequence was carried out according to a modification of the strategy outlined by Woyke et al. (2006) and extensively hand-curated to ensure validity of the bins. Further details of these procedures are given in the Supplementary Information.
Phylogenetic analysis of the 16S rRNA gene
Bacterial 16S rRNA genes were amplified with PCR primers 27f (5′-AGRGTTTGATCMTGGCTCAG-3′) and 1492r (5′-TACGGYTACCTTGTTAYGACTT-3′), cloned and sequenced from the DNA of samples BBAY01, 02, 04 and 15 as described by Shaw et al. (2008). In addition, a 16S rRNA gene library was produced for another sponge sample of T2 (named BBAY14). Mate pairs of each clone insert were assembled and sequences were checked for chimerae using Bellerophon (Huber et al., 2004). Only 16S rRNA gene sequences longer than 1200 bp were subsequently aligned to sequences in the Greengenes database (DeSantis et al., 2006) and their phylogeny was analysed using the maximum parsimony algorithm implemented in ARB (Ludwig et al., 2004). Clusters (at least 10 sequences with >99% identity) were identified and representative sequences were further analysed using the maximum likelihood algorithm implemented in ARB. For samples BBAY01, 02, 04, 14 and 15, the number of filtered sequences analysed was 771, 826, 709, 653 and 589, respectively.
Overlap between the 16S rRNA gene sequence in the sponge (n=1981) and the plankton (n=1597) was calculated by pairwise blastN comparison with a cutoff of 99% sequence identity and at least 97% common sequence coverage.
Results and discussion
Bacterial communities in C. concentrica and seawater are phylogenetically and functionally distinct
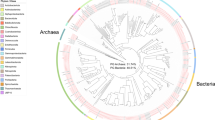
To further define the phylogenetic difference between the bacterial community of C. concentrica and the surrounding seawater, we generated 3545 16S rRNA gene sequences (>1200 bp) of replicate samples (Figure 1). Phylogenetic analysis showed that both bacterial communities contained less than 1% overlap (at a 99% identity cutoff). Specific differences are highlighted in this study with seawater communities harbouring members of the SAR11/Pelagibacter clade, OM1, Cytophaga and Flavobacteriales, bacterial groups commonly described in planktonic samples from around the globe (Giovannoni and Stingl, 2005). In contrast, the community of C. concentrica was dominated by distinct phylogenetic clusters within the γ-proteobacteria, Phyllobacteriaceae, Sphingomondales, Neisseriales and Nitrospiracae, among others (Figure 1). In particular, the α- and γ-proteobacterial groups are related to bacteria found in other sponges, as previously reported (Taylor et al., 2005, 2007b), whereas phyla such as Acidobacteria and Chloroflexi described to be present in some sponge species are absent in C. concentrica. A total of 34 sequences (0.95% of all sequences) were common to both the C. concentrica and the planktonic community. Twenty of those sequences were part of abundant planktonic sequence clusters shown in Figure 1 (Methylophilales, Roseobacter, Marinosulfonomonas (3x), Pelagibacter (3x), OM38_1 (3x), OM38_2, OM83_3, Cytophaga_1 (2x), Cytophaga_2 (3x), Cytophaga_4 (2 x) Cytophaga_5 (4 x) and OM1) and could be part of seawater not completely removed during the washing of the sponge (see Materials and methods). The remaining 24 sequences were not part of the abundant sponge clusters (Figure 1), only occurred once in either of the three sponge libraries and were related to single-count sequences in the planktonic libraries. These sequences might be allochthonous phylotypes shared with low-abundance populations in the plankton.
Phylogenetic comparison and abundance of dominant 16S RNA gene clusters of C. concentrica (light grey and marked with ‘C’) and surrounding seawater (dark grey and marked with ‘P’) bacterial communities. The tree on the left is based on the maximum likelihood analysis of a representative of each 16S rRNA gene cluster with at least 10 sequences. Clusters are named on the basis of the lowest taxonomic level that could be assigned using the Greengenes taxonomy (e.g., sponge cluster γ-proteobacteria_1 could not be classified below class level, whereas the cluster Piscirickettsiaceae is a cluster within the family Piscirickettsiaceae (class γ-proteobacteria)). Underscores followed by numbers in the name indicate that multiple distinct clusters (>99% identity) were found at a given taxonomic level. The histograms on the right represent the percentage of sequences assigned to clusters and the error bars indicate calculated standard variations of replicates (n=3 for C. concentrica, i.e., BBAY04, 14 and 15, and n=2 for seawater, i.e., BAY01 and 02; see Materials and methods). Clusters presented here contain 89.5 and 83.5% of the total 16S rRNA gene sequence recovered for the sponge (total 1951) and seawater samples (total 1597), respectively.
We estimate that the typical bacterial population of C. concentrica (3.82 × 108 bacterial cells per gram contained within the sponge's mesophyl and internal channel structure (Taylor et al., 2004a)) is exposed every day to planktonic cells approximately one order of magnitude greater in number (approximately 2.4 × 109 cells per gram per day), assuming that typical bacterial cell densities in seawater are around 105 cells per ml (Giovannoni and Stingl, 2005) and that up to 24 l of seawater can be filtered by a gram of sponge biomass per day (Vogel, 1977). Extensive culturing efforts in our laboratory and comparison of sponge sequences with 16S rRNA gene sequences from isolates in the Ribosomal Database Project (Cole et al., 2007) reveal that the sponge-associated bacteria detected in this study are not readily cultivated.
Random shotgun-sequencing data of replicate samples for the bacterial community of C. concentrica (92.6 Mbp unique sequence) and the surrounding seawater (67 Mbp unique sequence) were generated and assembled (Supplementary Table S1). Some of the larger scaffolds (>20 Kb) contained full-length 16S rRNA genes belonging to C. concentrica bacterial clusters in γ-proteobacteria 1, Phyllobacteriaceae, Sphingomondales, Piscirickettsiaceae and Bdellovibrionales. Compositional clustering of scaffolds supplemented with careful hand-curation and taxonomic assignment allowed for the recovery of partial genomes for these five new and uncultured organisms (Supplementary Figure S1). The taxonomic and functional gene composition of the entire community was assessed using an environmental gene tag analysis of assembled sequences (Tringe et al., 2005). The vast majority (>85%) of predicted and classifiable proteins could be taxonomically assigned to the domain bacteria, with less than 1.6% predicted to belong to archaeal organisms (Supplementary Table S1). This further suggests that the microbial community of C. concentrica is dominated by bacteria, in contrast to some other sponges that have been shown to possess high numbers of archaea (e.g. Preston et al., 1996).
Counts of functional gene annotations were adjusted for the relative coverage of the metagenome assembly, normalized for the number of genomes present in each sample and compared using statistical analysis of replicates. Overall functional comparison based on COG categories showed statistically significant differences in the key cellular biogenesis and metabolic processes of translation, ribosomal structure and biogenesis, secondary metabolite biosynthesis, transport and catabolism, replication, recombination and repair, nucleotide transport and metabolism, as well as energy production and conversion (Figure 2). Sponges have been noted for their diverse secondary metabolites (Sipkema et al., 2005), however, within the COG category ‘secondary metabolite biosynthesis, transport and catabolism’ none of the typical proteins associated with secondary metabolite biosynthesis, such as non-ribosomal peptide synthetases or polyketide synthetase, were abundant (less than three counts) in the C. concentrica data set, or significantly different in number to such proteins in the planktonic metagenome. As there is no information available on the secondary metabolite chemistry of C. concentrica, a correlation between the non-ribosomal peptide synthetases and polyketide synthetase identified in the data set and chemical compounds cannot be established.
Mobile genetic elements and genetic transfer
The overrepresentation of the COG category replication, recombination and repair in the metagenome of the sponge-associated bacteria is due to high numbers of transposable insertion elements (Figures 2 and 3). Large numbers of mobile elements have previously been noted in the genomes of symbiotic bacteria (Wu et al., 2004) and are proposed to have a crucial role in the evolution of bacterial genomes for symbiotic relationships with their hosts, for example, by disrupting non-required genes or by gene re-arrangements to generate new, required regulatory structures or pathways (Moran and Plague, 2004). Specifically, six COGs for transposases were more abundant in the sponge data set (by factors of 14–213) when compared with the planktonic data set (Figure 3 and Supplementary Table S2). In addition, the sponge metagenome had 2732 proteins (or an average of 64 per genome equivalent) with similarity to 378 different entries in a hand-curated database of insertion elements (Siguier et al., 2006). In contrast, only 47 proteins with similarity to 34 different elements were found in the planktonic data set (Supplementary Figure S2).
Abundance of specific COGs in sponge-associated and planktonic bacterial communities. COG counts were normalized per genome equivalent and all COGs shown are statistically different between the sponge and planktonic data set (P<0.05 in t-test). Error bars indicate calculated standard variations of replicates. Only COGs referred to in the text are presented; a complete list of all significant different COGs is given in Supplementary Table S2.
Specific sponge bacteria (such as the one assigned to Sphingomondales) contain one or more specific transposase types and seem to share transposase types with other members of the community (Supplementary Figure S1). In contrast, only two types of transposable insertion elements (with similarity to ISSsp2 and ISSde12) were found with greater than two counts in both the sponge-associated and planktonic data set (Supplementary Figure S2), indicating that there might be limited transfer of these genetic elements between the two bacterial communities. Two additional observations support this model of a genetic barrier. First, the sponge metagenome contains a relatively higher number of COG0610 (restriction enzymes type I helicase) and COG1715 (restriction endonuclease), which are both part of specific DNA modification and restriction systems. These two features seem to be shared between different members of the sponge community (Supplementary Figure S1) and might hence facilitate compatibility and exchange of DNA. In contrast, these DNA modification and restriction systems could function as a defence mechanism against foreign DNA derived from outside the sponge's bacterial community, which would help to maintain the integrity of the existing sponge–bacteria interaction. Second, a 66-fold higher frequency of clustered regularly interspaced short palindromic repeats (CRISPRs; see Supplementary Figure S3) and an overrepresentation of 10 TIGRFAM models describing CRISPR-associated proteins were observed in the sponge data set when compared with the planktonic metagenome (Supplementary Table S2). Together, CRISPR-associated proteins and CRISPRs form a viral-specific defence system (Barrangou et al., 2007). Virus titres in the ocean are typically around 107 per ml (Suttle, 2005), and given the water pumping rate of sponges (see above), we estimate that sponge-associated bacteria will be exposed to approximately 1000 viral particles per bacterial cell per day. An effective protection against bacteriolytic viral infection might therefore be an essential requirement for the survival of high cell-density, non-mobile communities, in which viruses would quickly spread and hence cause dramatic impact on bacterial population size. We note that these same mechanisms could maintain a barrier against phage-mediated gene transfer from the surrounding planktonic community, thereby also explaining the lack of overlap of transposable elements between sponge and planktonic communities.
Metabolism and stress response
The characteristics described above facilitate the persistence of a distinct bacterial community in C. concentrica, allowing for the maintenance of specific sponge–bacteria functional relationships. Sponge-associated bacteria have long been speculated to have evolved metabolic dependencies on their host or specific metabolic properties suitable for their environment (reviewed in Taylor et al., 2007b). Four observations from our analyses support this:
First, an overrepresentation of two key enzymes of the vitamin B12 synthesis pathways, uroporphyrinogen-III synthase (COG1587) and ATP:corrinoid adenosyltransferase/Cob(I)alamin adenosyltransferase (COG2109), indicates that members of the C. concentrica bacterial community in the sponge produce this essential vitamin (Figure 3). In particular, these enzymes were also found to be present in the sponge bacteria affiliated with Sphingomondales and Piscirickettsiaceae (Supplementary Figure S1). Eukaryotes have so far not been reported to produce vitamin B12 and need to acquire this essential cofactor through food (Roth et al., 1996). It has also been postulated that vitamin B12 is acquired by some macroalgal species through transfer from surface-associated, symbiotic bacteria (Croft et al., 2005). Our results also imply that vitamin B12 dependency in the sponge could be satisfied by resident bacterial community members rather than by food bacteria filtered from the plankton.
Second, bacterial communities have recently been shown to nitrify the ammonium that accumulates and is being secreted by the sponge tissue of Aplysina aerophoba (Bayer et al., 2008). Highly regulated ammonium assimilation in the bacterial community of C. concentrica is indicated by the abundance of nitrogen regulatory protein PII (COG0347), which controls the transcription of the glutamine synthase gene (glnA) on the basis of the ratio of glutamine to 2-ketoglutarate, and by glutamate dehydrogenase (COG2902), which catalyses the reversible deamination of L-glutamate to 2-ketoglutarate (Figure 3). The nitrogen regulatory protein PII even occurs in multiple copies in the partial genome of Phyllobacteriaceae and γ-protobacteria 1 groups (Supplementary Figure S1). In contrast, asparagine synthase (glutamine-hydrolyzing AsnB; COG0367), which catalyses the formation of asparagine using either ammonium or glutamine as amide donor, was found more frequently (by a factor of 12) in the planktonic community. This suggests that specific pathways and regulation of assimilation are preferred in the sponge-associated community and that assimilation processes rather than oxidation might be important for the ammonium utilization by the bacterial community in C. concentrica. In addition, anaerobic pathways such as sulphate reduction, denitrification or anammox, which have been described in deep-water sponge systems (Hoffmann et al., 2009; Bruck et al., 2010), are not abundant in the C. concentrica metagenome, which is consistent with the thin structure of this shallow-water sponge's being unlikely to experience anaerobiosis.
Third, free-living, planktonic bacteria are expected to encounter types and amounts of nutrients and ions, different from those experienced by bacteria associated with a host. This is reflected in differences in the classes of transport systems found in the two data sets, with COG 1462 (permease component of an ABC-transporter), COG 3090 (TRAP-type C4-dicarboxylate transport system, small permease component), COG 0025 (NhaP-type Na+/H+ and K+/H+ antiporters) and COG 2223 (Nitrate/nitrite transporter) being overrepresented and COG0659 (Sulphate permease and related transporters, major facilitator superfamily) being underrepresented in the sponge's bacterial community (Figure 3).
Fourth, the large (COG1529) and middle (COG1319) subunits of the aerobic-type carbon monoxide dehydrogenase were overrepresented by a factor of greater than three in the sponge metagenome (Figure 3; the small subunit (COG2080) is more abundant by a factor of 2.2, t-test, P=0.047), indicating that a portion of the sponge bacterial community can generate reductive energy from the oxidation of CO. Further analysis revealed that both form I and putative form II carbon monoxide dehydrogenase are present (King and Weber, 2007), with no apparent preference in the sponge or plankton data set (Supplementary Figure S4). Lithoheterotrophy based on CO has only been recently recognized in marine bacterioplankton (Moran and Miller, 2007), and in this study, we present evidence for a presence of this process in sponge bacteria. In particular, the carbon monoxide dehydrogenase function could be assigned to the Phyllobacteriaceae and Sphingomondales groups (Supplementary Figure S1), with the latter order taxon not previously reported to possess the potential for CO oxidation activity.
Bacteria associated with a sponge are also likely to experience greater temporal variation in their environment, relative to planktonic bacteria, depending on the extent of water pumping, the metabolic rate or the conditions of the host or other environmental factors (Vogel, 1977). Thus, the availability of electron acceptors and nutrients may fluctuate substantially and the abundance of peroxiredoxin (COG0450) and glutathione-S-transferases (COG0625) might be important factors to control cytoplasmic redox balance and oxidative damage caused by redox-active by-products under these conditions (Vuilleumier, 1997; Hofmann et al., 2002). The sponge metagenome also contains a high count of coenzyme F390 synthetase (COG1541), an enzyme that converts factor F420 to coenzyme F390 under aerobic conditions. Coenzyme F390 has so far only been reported for methanogens and has been suggested to function as a redox reporter and response regulator (Vermeij et al., 1997). Other genomic signatures for methanogens were absent in the sponge data set, indicating that the role of coenzyme F390 synthetase might not be restricted to methanogens and could be an adaptive feature for the redox response of sponge-associated bacteria. In fact, in two cases (Phyllobacteriaceae and Sphingomondales, Supplementary Figure S1), the presence of coenzyme F390 synthetase could be directly linked to a bacterial origin.
Bacteria–sponge interaction through repeat proteins
We observed a significant and substantial overrepresentation of ankyrin repeat (AR) (by a factor of 246±35) and TPR (by a factor of 136±24) proteins for the sponge bacterial community (Figure 3). TPR and AR mediate protein–protein interactions in eukaryotes and have been found in proteins involved in various functional processes, including transcriptional initiators, cell cycle regulators, cytoskeleton proteins, ion transporters and signal transducers (Blatch and Lassle, 1999; Hryniewicz-Jankowska et al., 2002). Taxonomic assignment of the sponge community-derived sequences excluded them from being derived from potentially contaminating eukaryotes (see Material and Methods). Further analysis of all AR and TPR proteins against the Pfam database supported their annotation and showed that up to 37 repeats per protein could be found (median 6 and 3 for AR and TPR proteins, respectively; Supplementary Figure S5). Phylogenetic analysis of the secreted AR proteins showed clustering with groups of predominantly bacterial sequences, including the insect endosymbiont Wolbachia (Supplementary Figure S4). A total of 20 AR and 13 TPR proteins could be directly linked to four of the five sponge-bacterial groups (Supplementary Figure S1) and genomic clustering into ‘AR/TPR islands’ was observed in two cases (see Figure 4). The abundance of AR proteins has been noted in the genomes of obligate and facultative intracellular symbionts of eukaryotic cells (Wu et al., 2004). For the sponge data set, 26 out of 121 (20.7%) and 14 out of 66 (21.1%) of the AR and TPR proteins, respectively, have signal peptides for extracellular secretion in Gram-negative bacteria. This suggests that those proteins have functions outside the bacterial cytoplasm and most likely interact with surrounding cells and their proteins. AR proteins have also been recently shown to be secreted by the intracellular pathogens Legionella pneumophila and Coxiella burnetii and to interfere with the microtubule-dependent vascular transport of host cells, hence blocking the normal progression of phagocytosis (Pan et al., 2008). As uptake of food bacteria by sponge cells is also mediated by phagocytosis (Wehrl et al., 2007), the sponge-specific AR and TPR proteins reported in this study could possibly represent a mechanism by which symbiotic bacteria avoid digestion, and could explain the long-standing question of how food and symbionts are discriminated by the sponge (Wilkinson et al., 1984).
Clustering of AR and TPR proteins in the genome of two uncultured sponge-associated bacteria belonging to the Bdellovibrionales order and Phyllobacteriaceae family. Light grey arrows represent ORFs and dark grey boxes highlight ‘AR/TPR islands’. Relative positions on the genomic scaffolds are given.
Conclusion
The specific, genomic signatures identified in this study have hitherto not been recognized to be involved in mediating the interactions between bacteria and their sponge host or within the bacterial community under the particular biological, chemical and physical conditions provided by the sponge environment. Thus, those signatures provide insight into the potentially specific mechanisms by which distinct bacteria persist in the sponge in the face of the flood of food bacteria entering the system from the plankton, the metabolic interdependencies of the two partners and how the relationship might be maintained as a mutualism. Our data set also delivers novel markers for monitoring the status of the sponge bacterial community and hence assesses how abundance, diversity or expression of those functional genes affect the symbiotic relationship between bacteria and sponges. This is particularly important, as phylogenetic shifts in the bacterial community composition have been correlated with sponge disease (Webster, 2007), and in at least one case this has been linked to temperature changes (Webster et al., 2008). Yet, the functional changes underpinning the breakdown of symbiosis are completely unexplored.
References
Altschul SF, Gish W, Miller W, Myers EF, Lipman DJ . (1990). Basic local alignment search tool. J Mol Biol 215: 403–410.
Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712.
Bayer K, Schmitt S, Hentschel U . (2008). Physiology, phylogeny and in situ evidence for bacterial and archaeal nitrifiers in the marine sponge Aplysina aerophoba. Environ Microbiol 10: 2942–2955.
Bendtsen JD, Nielsen H, von Heijne G, Brunak S . (2004). Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340: 783–795.
Blatch GL, Lassle M . (1999). The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21: 932–939.
Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E et al. (2003). The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res 31: 365–370.
Bruck WM, Bruck TB, Self WT, Reed JK, Nitecki SS, McCarthy PJ . (2010). Comparison of the anaerobic microbiota of deep-water Geodia spp. and sandy sediments in the Straits of Florida. ISME J 4: 686–699.
Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P . (2006). Toward automatic reconstruction of a highly resolved tree of life. Science 311: 1283–1287.
Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM et al. (2007). The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res 35: D169–D172.
Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG . (2005). Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438: 90–93.
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072.
Eddy SR . (1998). Profile hidden Markov models. Bioinformatics 14: 755–763.
Fieseler L, Quaiser A, Schleper C, Hentschel U . (2006). Analysis of the first genome fragment from the marine sponge-associated, novel candidate phylum Poribacteria by environmental genomics. Environ Microbiol 8: 612–624.
Giovannoni SJ, Stingl U . (2005). Molecular diversity and ecology of microbial plankton. Nature 437: 343–348.
Grissa I, Vergnaud G, Pourcel C . (2007). The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8: 172.
Haft DH, Selengut JD, White O . (2003). The TIGRFAMs database of protein families. Nucleic Acids Res 31: 371–373.
Hoffmann F, Radax R, Woebken D, Holtappels M, Lavik G, Rapp HT et al. (2009). Complex nitrogen cycling in the sponge Geodia barretti. Environ Microbiol 11: 2228–2243.
Hofmann B, Hecht HJ, Flohe L . (2002). Peroxiredoxins. Biol Chem 383: 347–364.
Hryniewicz-Jankowska A, Czogalla A, Bok E, Sikorsk AF . (2002). Ankyrins, multifunctional proteins involved in many cellular pathways. Folia Histochem Cytobiol 40: 239–249.
Huber T, Faulkner G, Hugenholtz P . (2004). Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20: 2317–2319.
Huson DH, Auch AF, Qi J, Schuster SC . (2007). MEGAN analysis of metagenomic data. Genome Res 17: 377–386.
King GM, Weber CF . (2007). Distribution, diversity and ecology of aerobic CO-oxidizing bacteria. Nat Rev Microbiol 5: 107–118.
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371.
Moran MA, Miller WL . (2007). Resourceful heterotrophs make the most of light in the coastal ocean. Nat Rev Microbiol 5: 792–800.
Moran NA, Plague GR . (2004). Genomic changes following host restriction in bacteria. Curr Opin Genet Dev 14: 627–633.
Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, Flanigan MJ et al. (2000). A whole-genome assembly of Drosophila. Science 287: 2196–2204.
Noguchi H, Park J, Takagi T . (2006). MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res 34: 5623–5630.
Pan X, Luhrmann A, Satoh A, Laskowski-Arce MA, Roy CR . (2008). Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 320: 1651–1654.
Preston CM, Wu KY, Molinski TF, DeLong EF . (1996). A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov., sp. nov. Proc Natl Acad Sci USA 93: 6241–6246.
Roth JR, Lawrence JG, Bobik TA . (1996). Cobalamin (coenzyme B12): synthesis and biological significance. Annu Rev Microbiol 50: 137–181.
Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S et al. (2007). The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol 5: e77.
Sammut SJ, Finn RD, Bateman A . (2008). Pfam 10 years on: 10 000 families and still growing. Brief Bioinform 9: 210–219.
Schirmer A, Gadkari R, Reeves CD, Ibrahim F, DeLong EF, Hutchinson CR . (2005). Metagenomic analysis reveals diverse polyketide synthase gene clusters in microorganisms associated with the marine sponge Discodermia dissoluta. Appl Environ Microbiol 71: 4840–4849.
Shaw AK, Halpern AL, Beeson K, Tran B, Venter JC, Martiny JB . (2008). It's all relative: ranking the diversity of aquatic bacterial communities. Environ Microbiol 10: 2200–2210.
Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M . (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34: D32–D36.
Sipkema D, Franssen MC, Osinga R, Tramper J, Wijffels RH . (2005). Marine sponges as pharmacy. Mar Biotechnol (NY) 7: 142–162.
Suttle CA . (2005). Viruses in the sea. Nature 437: 356–361.
Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV et al. (2003). The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4: 41.
Taylor MW, Baillie HJ, Charlton TS, De Nys R, Kjelleberg S, Steinberg PD . (2004a). AHL production in bacteria associated with marine benthic eukaryotes. Appl Environ Microbiol 4387-9: 87–89.
Taylor MW, Hill RT, Piel J, Thacker RW, Hentschel U . (2007a). Soaking it up: the complex lives of marine sponges and their microbial associates. ISME J 1: 187–190.
Taylor MW, Radax R, Steger D, Wagner M . (2007b). Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol Mol Biol Rev 71: 295–347.
Taylor MW, Schupp PJ, Dahllof I, Kjelleberg S, Steinberg PD . (2004b). Host specificity in marine sponge-associated bacteria, and potential implications for marine microbial diversity. Environ Microbiol 6: 121–130.
Taylor MW, Schupp PJ, de Nys R, Kjelleberg S, Steinberg PD . (2005). Biogeography of bacteria associated with the marine sponge Cymbastela concentrica. Environ Microbiol 7: 419–433.
Thompson JD, Gibson TJ, Higgins DG . (2002). Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Chapter 2: Unit 2 3.
Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW et al. (2005). Comparative metagenomics of microbial communities. Science 308: 554–557.
Tringe SG, Zhang T, Liu X, Yu Y, Lee WH, Yap J et al. (2008). The airborne metagenome in an indoor urban environment. PLoS ONE 3: e1862.
Vermeij P, Pennings JL, Maassen SM, Keltjens JT, Vogels GD . (1997). Cellular levels of factor 390 and methanogenic enzymes during growth of Methanobacterium thermoautotrophicum deltaH. J Bacteriol 179: 6640–6648.
Vogel G . (2008). The inner lives of sponges. Science 320: 1028–1030.
Vogel S . (1977). Current-induced flow through living sponges in nature. Proc Natl Acad Sci USA 74: 2069–2071.
Vuilleumier S . (1997). Bacterial glutathione S-transferases: what are they good for? J Bacteriol 179: 1431–1441.
Webster NS . (2007). Sponge disease: a global threat? Environ Microbiol 9: 1363–1375.
Webster NS, Blackall LL . (2008). What do we really know about sponge-microbial symbioses? ISME J 3: 1–3.
Webster NS, Cobb RE, Negri AP . (2008). Temperature thresholds for bacterial symbiosis with a sponge. ISME J 2: 830–842.
Wehrl M, Steinert M, Hentschel U . (2007). Bacterial uptake by the marine sponge Aplysina aerophoba. Microb Ecol 53: 355–365.
Wilkinson C, Garrone G, Vacelet J . (1984). Marine sponges discriminate between food bacteria and bacterial symbionts: electron microscope radioautography and in situ evidence. Proc R Soc London, B 220: 519–528.
Woyke T, Teeling H, Ivanova NN, Huntemann M, Richter M, Gloeckner FO et al. (2006). Symbiosis insights through metagenomic analysis of a microbial consortium. Nature 443: 950–955.
Wu M, Sun LV, Vamathevan J, Riegler M, Deboy R, Brownlie JC et al. (2004). Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol 2: E69.
Acknowledgements
We thank Mike Chandler (Laboratoire de Microbiologie et Genetique Moleculaires, Toulouse Cedex, France) for access to the hand-curated database of IS elements, and Sharon Longford and Rick Cavicchioli for their help during field sampling. We thank Granger Sutton and his team for the ongoing development and maintenance of the Celera Assembler. We thank Robert Friedman and acknowledge the J Craig Venter Institute (JCVI) Joint Technology Center, under the leadership of Yu-Hui Rogers, for producing the genomic libraries and sequence data. This work was supported by funds from the Gordon and Betty Moore Foundation and the Australian Research Council (LP0668235). The shotgun sequencing and 16S rRNA gene data of this project have been deposited at GenBank under GenomeProject ID 34751 and are also available through the Community Cyberinfrastructure for Advanced Marine Microbial Ecology Research and Analysis (CAMERA) website (http://camera.calit2.net/).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Thomas, T., Rusch, D., DeMaere, M. et al. Functional genomic signatures of sponge bacteria reveal unique and shared features of symbiosis. ISME J 4, 1557–1567 (2010). https://doi.org/10.1038/ismej.2010.74
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2010.74
Keywords
This article is cited by
-
Bacterial aerobic methane cycling by the marine sponge-associated microbiome
Microbiome (2023)
-
Microbiome species diversity and seasonal stability of two temperate marine sponges Hymeniacidon perlevis and Suberites massa
Environmental Microbiome (2023)
-
Microbiome profile of the Antarctic clam Laternula elliptica
Brazilian Journal of Microbiology (2023)
-
Symbiont transmission in marine sponges: reproduction, development, and metamorphosis
BMC Biology (2022)
-
Microbiome diversity and metabolic capacity determines the trophic ecology of the holobiont in Caribbean sponges
ISME Communications (2022)