Abstract
One of the main tasks of NASA's planetary protection program is to prevent the forward contamination of extraterrestrial environments with Earth life, and in turn preserve other planets and the integrity of future life detection missions. Despite information regarding bacterial diversity in NASA's clean rooms, little is known about the presence of Archaea. Archaeal community analysis of spacecraft-associated surfaces is important, as they are considered by some to represent terrestrial life most capable of surviving on Mars. The first insights into the archaeal diversity of clean rooms where spacecraft assembled are attempted. Nucleic acid sequences clustering with uncultivable Archaea within the Eury- and Crenarchaeota were retrieved from 8 of 26 samples collected from several spacecraft assembly clean rooms. Due to their potential capability to survive and proliferate in Martian conditions, screening for Archaea on spacecraft surfaces and instruments that are associated with future life detection missions may be necessary.
Similar content being viewed by others

Main
The presence of non-extremophilic Archaea in most biological niches (Olsen, 1994; Bintrim et al., 1997; Schouten et al., 2000) coupled with the varied metabolisms of Archaea as a whole suggests that these microbes play a significant role in Earth's ecology (DeLong et al., 1994). Due to this ubiquity and metabolic diversity, Archaea are considered by some to be capable of surviving, or even thriving, on Mars (Krasnopolsky, 2006). The potential discovery of (ancient) liquid water on Mars (Herkenhoff et al., 2004) and the chemical compositions of the Martian atmosphere (Weiss et al., 2000) and crust (Fisk and Giovannoni, 1999) not only raises the possibility that life may have or may still exist there, but indicates that conditions capable of supporting terrestrial life may also be present. The prevention of contamination of Mars and other extraterrestrial environments with terrestrial biomolecules and/or life (Rummel, 1989) is therefore tremendously important to avoid confounding future life detection missions and preserve the pristine conditions of other solar bodies.
Current NASA planetary protection protocols for determining microbial burden on spacecraft surfaces were developed using the detection of aerobic spore-forming bacteria (NASA, 2005). The recent isolation of extremotolerant bacteria (La Duc et al., 2007) and the presence of viable but yet to be cultivated bacteria (Moissl et al., 2007) from various spacecraft assembly facility (SAF) clean rooms suggested the need to evaluate other potential microbial communities. Therefore, samples collected from spacecraft-associated clean room facilities at the Jet Propulsion Laboratory (JPL) and Johnson Space Center (JSC) were evaluated for archaeal signatures.
In total, 26 samples were collected (18 from JPL and 8 from JSC; Table 1) and subjected to DNA extraction (Moissl et al., 2007) and Archaea-specific PCR amplification for ∼1100 bp fragments using appropriate primer sets (methods given in Supplementary Information). Despite strict NASA clean room conditions (NASA-KSC, 1999) and demonstrated low microbial bioburden (La Duc et al., 2007), 6 of 18 JPL-SAF and 2 of 8 JSC-Genesis Curation Laboratory (GCL) samples exhibited positive archaeal 16S rRNA gene amplification (Table 1). Air samples were collected from outside the facility as described elsewhere (Moissl et al., 2007), however the archaeal specific fragments were not obtained. Attempts to cultivate Archaea from clean room samples or to obtain positive archaeal signals using fluorescent in situ hybridization were unsuccessful. Clones selected for sequencing, and the RFLP patterns and operational taxonomic units (OTU) obtained are given in Table 1. Rarefaction analyses indicated the coverage value to be 100% since none of the OTU's retrieved from any of the tested samples appeared only once.
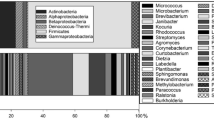
The results suggested that both facilities contained a wide diversity of Archaea (Figure 1). Among 542 clones analyzed, 31 different archaeal sequences were obtained: 26 from the JPL-SAF (479 clones) and 5 from JSC-GCL (63 clones) locations. In general, individual archaeal sequences were unique to a given facility or location. The majority of sequences were affiliated with an uncultivable soil Crenarchaeota group (Figure 1). Clones showed sequence differences up to 7.8% with the nearest identified archaeal species, and clustered with sequences obtained from deep South African gold mine waters (Takai et al., 2001), soil (Bintrim et al., 1997), rhizospheres (Simon et al., 2005), permafrost soils (Ochsenreiter et al., 2003) and the leachate of landfill (Laloui-Carpentier et al., 2006). Clones obtained were not closely related to cultivable Archaea; the 16S rRNA gene sequence of the nearest cultured neighbor, the autotrophic ammonia-oxidizing crenarchaeon candidatus Nitrosopumilus maritimus (Konneke et al., 2005), was up to 16% different.
Phylogenetic tree (Maximum Parsimony), showing the archaeal diversity and the phylogenetic affiliation of the clone sequences derived from different sampling locations. The GenBank accession numbers of the clones retrieved from this study (prefix DQ) and reference strains are given. The facility ID are SAF, JPL-SAF; JSC, JSC-GCL. The clones are identified by the facility ID (SAF or JSC) preceded by the month of collection (SAF only) and followed by the location and representative clone number (i.e., 3SAF2-2 represents clone 2 from location 2 collected from the JPL-SAF during March). The numbers in brackets give the sequence length. The scale bar shows a 10% estimated difference in nucleotide sequence positions.
Additionally, a single clone sequence (ARC_3SAF2-2, Figure 1) from JPL-SAF was affiliated with methanogens in the Euryarchaeal phylum. The sequence clustered with 16S rRNA gene sequences from uncultivable Archaea present in temperate anoxic soils (Wu et al., 2006) showed at least 18% sequence difference to the closest cultivable neighbor. Although methanogens have generally been regarded as strict anaerobes, it was demonstrated that some methanogens were able to withstand long exposure to high levels of oxygen (Kato et al., 1997) and therefore the aerobic conditions of the clean room may not necessarily be lethal.
The absence of archaeal signatures from two-thirds of the collected samples (and all of the air samples) demonstrated the relative sterility of these clean rooms. A lack of similar sequences between the certified and uncertified rooms (JSC7 and JSC9, respectively) further illustrated the effectiveness of the quality control protocols enforced to maintain these facilities (NASA-KSC, 1999).
All retrieved sequences belonged to the Crenarchaeota and Euryarcheota phyla. This may be due in part to the primer selection as no truly universal Archaeal primer sets have been established and it was unlikely that any Nanoarchaetoa sequences would have been amplified given the primers utilized.
All of the archaeal sequences retrieved during this study belonged to clusters of uncultivable Archaea from cold or moderate environments and their structural properties and physiological capabilities remain unknown. It was unclear whether the detected Archaea were alive at the time of sampling, would be capable of growth in extraterrestrial environments, or able to withstand space travel, but given the ubiquitous nature of Archaea it would be naive to completely dismiss any of these possibilities. The detection of Archaea in these controlled sanitized environments further emphasized the potential of these microbes to exist in the most extreme biotopes. Future studies utilizing quantitative detection methods as well as the verification of archaeal viability could lead to better characterization and enable the eradication of these microbes.
To date, as per international treaty, it is required to estimate number of spores on spacecraft surfaces using conventional culture-based method (NASA, 2005). While this may be a valid proxy for overall spacecraft cleanliness, this study has clearly demonstrated the presence of Archaea on different surfaces of these spacecraft-associated clean rooms. As Archaea may be considered terrestrial organisms capable of living or surviving on Mars and other extraterrestrial environments, the screening for these microbes on spacecraft surfaces and instruments associated with future life detection missions may be necessary to maintain mission integrity and prevent forward contamination.
References
Bintrim SB, Donohue TJ, Handelsman J, Roberts GP, Goodman RM . (1997). Molecular phylogeny of Archaea from soil. Proc Natl Acad Sci USA 94: 277–282.
DeLong EF, Wu KY, Prezelin BB, Jovine RV . (1994). High abundance of Archaea in Antarctic marine picoplankton. Nature 371: 695–697.
Fisk MR, Giovannoni SJ . (1999). Sources of nutrients and energy for a deep biosphere on Mars. J Geophys Res 104: 11805–11816.
Herkenhoff KE, Squyres SW, Arvidson R, Bass DS, Bell JF, Bertelsen P et al. (2004). Evidence from opportunity's microscopic imager for water on Meridiani Planum. Science 306: 1727–1730.
Kato MT, Field JA, Lettinga G . (1997). Anaerobe tolerance to oxygen and the potentials of anaerobic and aerobic cocultures for wastewater treatment. Braz J Chem Eng 14 (Available from:〈http://www.scielo.br/scielo.php?script=sci_arttext&pid=S0104-66321997000400015&lng=en&nrm=iso〉).
Konneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA . (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437: 543–546.
Krasnopolsky VA . (2006). Some problems related to the origin of methane on Mars. Icarus 180: 359–367.
La Duc MT, Dekas AE, Osman S, Moissl C, Newcombe D, Venkateswaran K . (2007). Isolation and characterization of bacteria capable of tolerating the extreme conditions of clean-room environments. Appl Environ Microbiol 73: 2600–2611.
Laloui-Carpentier W, Li T, Vigneron V, Mazéas L, Bouchez T . (2006). Methanogenic diversity and activity in municipal solid waste landfill leachates. Antonie Van Leeuwenhoek 89: 423–434.
Moissl C, La Duc MT, Osman S, Dekas AE, Venkateswaran K . (2007). Molecular bacterial community analysis of clean rooms where spacecraft are assembled. FEMS Microbiol Ecol 61: 509–521.
NASA (2005). Planetary Protection Provisions for Robotic Extraterrestrial Missions. NPR 8020.12C, April 2005. National Aeronautics and Space Administration: Washington, DC.
NASA-KSC (1999). Launch Site Requirement Planning Group, Facilities Handbook of Payload Hazardous Servicing Facility (PHSF), K-STSM-14.1.15 rev D. Edited by NASA-KSC: KSC: Cape Canaveral, FL.
Ochsenreiter T, Selezi D, Quaiser A, Bonch-Osmolovskaya L, Schleper C . (2003). Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ Microbiol 5: 787–797.
Olsen GJ . (1994). Microbial ecology. Archaea, Archaea, everywhere. Nature 371: 657–658.
Rummel JD . (1989). Planetary protection policy overview and application to future missions. Adv Space Res 9: 181–184.
Schouten S, Hopmans EC, Pancost RD, Damste JS . (2000). Widespread occurrence of structurally diverse tetraether membrane lipids: evidence for the ubiquitous presence of low-temperature relatives of hyperthermophiles. Proc Natl Acad Sci USA 97: 14421–14426.
Simon HM, Jahn CE, Bergerud LT, Sliwinski MK, Weimer PJ, Willis DK et al. (2005). Cultivation of mesophilic soil crenarchaeotes in enrichment cultures from plant roots. Appl Environ Microbiol 71: 4751–4760.
Takai K, Moser DP, DeFlaun M, Onstott TC, Fredrickson JK . (2001). Archaeal diversity in waters from deep South African gold mines. Appl Environ Microbiol 67: 5750–5760.
Weiss BP, Yung YL, Nealson KH . (2000). Atmospheric energy for subsurface life on Mars? Proc Natl Acad Sci USA 97: 1395–1399.
Wu XL, Friedrich MW, Conrad R . (2006). Diversity and ubiquity of thermophilic methanogenic archaea in temperate anoxic soils. Environ Microbiol 8: 394–404.
Acknowledgements
This research was carried out at JPL/Caltech under a contract with NASA and funded by Mars Sample Return Mission program as well as by NRA ROSS 2005. We thank Jerome Smith and Anne Dekas (JPL), Judith Allton, Karen McNamara, and Carlton Allen (JSC) for helping to sample various locations and Shariff Osman for critically reading this manuscript. We acknowledge John Rummel, NASA Planetary Protection Officer for constant encouragements and Jason Kastner for facilitating various lab facilities.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary information
Rights and permissions
About this article
Cite this article
Moissl, C., Bruckner, J. & Venkateswaran, K. Archaeal diversity analysis of spacecraft assembly clean rooms. ISME J 2, 115–119 (2008). https://doi.org/10.1038/ismej.2007.98
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2007.98
Keywords
This article is cited by
-
Microscopic Characterization of Biological and Inert Particles Associated with Spacecraft Assembly Cleanroom
Scientific Reports (2019)
-
Advanced Curation of Astromaterials for Planetary Science
Space Science Reviews (2019)
-
Human age and skin physiology shape diversity and abundance of Archaea on skin
Scientific Reports (2017)
-
Microbial succession in an inflated lunar/Mars analog habitat during a 30-day human occupation
Microbiome (2016)
-
A viability-linked metagenomic analysis of cleanroom environments: eukarya, prokaryotes, and viruses
Microbiome (2015)