Abstract
Collectively, phagotrophic algae (mixotrophs) form a functional continuum of nutritional modes between autotrophy and heterotrophy, but the specific physiological benefits of mixotrophic nutrition differ among taxa. Ochromonas spp. are ubiquitous chrysophytes that exhibit high nutritional flexibility, although most species generally fall towards the heterotrophic end of the mixotrophy spectrum. We assessed the sources of carbon and nitrogen in Ochromonas sp. strain BG-1 growing mixotrophically via short-term stable isotope probing. An axenic culture was grown in the presence of either heat-killed bacteria enriched with 15N and 13C, or unlabeled heat-killed bacteria and labeled inorganic substrates (13C-bicarbonate and 15N-ammonium). The alga exhibited high growth rates (up to 2 divisions per day) only until heat-killed bacteria were depleted. NanoSIMS and bulk IRMS isotope analyses revealed that Ochromonas obtained 84–99% of its carbon and 88–95% of its nitrogen from consumed bacteria. The chrysophyte assimilated inorganic 13C-carbon and 15N-nitrogen when bacterial abundances were very low, but autotrophic (photosynthetic) activity was insufficient to support net population growth of the alga. Our use of nanoSIMS represents its first application towards the study of a mixotrophic alga, enabling a better understanding and quantitative assessment of carbon and nutrient acquisition by this species.
Similar content being viewed by others
Introduction
Aquatic microbial food webs encompass interactions between bacteria, cyanobacteria, phototrophic and heterotrophic protists, and viruses (Azam and Malfatti, 2007; Sarmento, 2012; Worden et al., 2015). Models of these food webs generally characterize eukaryotic microbes as either phototrophic (capable of producing energy and fixing carbon for growth using only inorganic compounds and light) or heterotrophic (dependent on the consumption of organic material for nutrition). These characterizations greatly oversimplify trophic interactions because many microbial eukaryotic algae are mixotrophic, capable of using both inorganic and organic forms of carbon and major nutrients (Flynn et al., 2013). Although the mixotrophic nature of many algae is now recognized, the impact of this behavior in terms of carbon and nutrient cycling, as well as its ecological significance for the algae that conduct this behavior, is not widely acknowledged or understood. Some attempts have been made to classify mixotrophs in a few conceptual models (Våge et al., 2013; Andersen et al., 2015; Mitra et al., 2016; Ward and Follows, 2016), but the importance of mixotrophic organisms to food web structure and function has not yet routinely been entrained into mathematical models of food web dynamics.
Mixotrophic species exhibit a range of nutritional behaviors. Some species are predominantly phototrophic but supplement photosynthesis with a small amount of heterotrophy, while other species are predominantly heterotrophic and use photosynthesis only for extending survival when prey are scarce (Jones, 1994, 2000; Rottberger et al., 2013). Mixed nutrition appears to provide an advantage particularly in oligotrophic ecosystems (Hartmann et al., 2012; Unrein et al., 2014), presumably because nutrients are often limiting to purely phototrophic protists, while strict heterotrophs are limited by prey availability. In contrast, mixotrophic protists may meet their elemental and energy demands through a mixture of photosynthesis and prey ingestion. Yet, mixotrophic algae do not dominate all ecosystems, and therefore mixed nutrition appears to impose tradeoffs that are not well understood (Raven, 1997; Stoecker, 1998).
More than 100 mixotrophic species of Chrysophyceae have been described, and sequence-based surveys of natural samples point to potentially larger diversity (Andersen, 2007; del Campo and Massana, 2011). The group presents interesting model species for investigating mixotrophy due to the wide diversity of nutritional strategies among these species. Some, such as Uroglena americana, appear to be primarily autotrophic but require a phospholipid that is obtained via phagocytosis of prey (Kimura and Ishida, 1989). A similar case has been reported for Dinobryon cylindricum, where phagocytosis might also provide essential growth factors and/or nutrients for photosynthetic growth (Caron et al., 1993). At the other end of the spectrum, species exist that exhibit primarily heterotrophic metabolism where phototrophy appears to be of minor significance (Andersson et al., 1989; Caron et al., 1990; Boraas et al., 1992; Sanders et al., 2001; Foster and Chrzanowski, 2012; Chrzanowski and Foster, 2014; Wilken et al., 2014).
Experiments with mixotrophic algae have yielded results that outline the overall metabolism of these organisms (Sanders and Porter, 1988; Sanders, 2011), but most studies have not clearly defined the sources (phototrophy or heterotrophy) of carbon and nutrients in these species. Similarly, recent molecular studies offer insights into the metabolic potential of these algae (Liu et al., 2015; Terrado et al., 2015; Liu et al., 2016), but do not specifically quantify the contributions of photosynthesis and heterotrophy to mixotrophic nutrition. Stable isotope probing (SIP) offers a method to address this issue. Combining SIP with high-resolution nanometer-scale secondary ion mass spectrometry (nanoSIMS), researchers have been able to quantify and map elemental and isotopic abundances at 50–100 nm resolution in order to assess the metabolic activities and elemental flow within individual cells (Boxer et al., 2009; Gao et al., 2016). The approach has been used to characterize the metabolic potential and anabolic activity in environmental microorganisms (Orphan and House, 2009; Orphan, 2009), including the utilization of different carbon substrates by deep sub-seafloor bacterial cells (Morono et al., 2011), carbon and nitrogen utilization by cyanobacteria (Popa et al., 2007; Finzi-Hart et al., 2009), the flux of nitrogen within coral symbionts (Kopp et al., 2013), the transfer of nitrogen from lysed algae to bacteria (Sheik et al., 2014) and the transfer of carbon to bacterial surface symbionts of a large cellulolytic protist symbiont of a lower termite (Carpenter et al., 2013).
Here we apply stable isotope probing and nanoSIMS to an axenic culture of the mixotrophic chrysophyte, Ochromonas sp. strain BG-1 (Sanders et al., 2001). Incubations were conducted with the alga in the presence of either labeled inorganic substrates or labeled heat-killed bacteria, in order to identify and quantify the sources of carbon and nitrogen utilized by this alga. Given its affinity for bacterial prey (Sanders et al., 2001), we hypothesize that this Ochromonas strain, in the presence of high bacterial abundances, would acquire an elevated percentage of its carbon and nitrogen requirements through heterotrophy via the ingestion and assimilation of bacteria. Our analysis showed that while defined as a mixotroph, this strain of Ochromonas relies primarily on bacterial phagotrophy for both carbon and nitrogen acquisition and that photosynthesis remains a minor aspect of its nutrition.
Materials and methods
Bacterial isolation and production of heat-killed bacteria
Heat-killed bacteria (HKB) were prepared as non-living prey for all experimental work in this study to avoid uptake or release of organic or inorganic substances by bacteria in the axenic culture of the chrysophyte. Bacteria from a bacterized culture of Ochromonas BG-1 were streaked on petri dishes with a solid agar medium containing a minimal M9 medium (Marley et al., 2001) using ammonium as the sole source of nitrogen and glucose as the sole source of carbon. Individual colonies were transferred to 25 ml tubes containing the M9 medium (without agar), which were used to inoculate 1.5 l of medium and incubated at 20 °C on a shaker table in order to obtain large numbers of bacteria for the production of HKB. Cultures were grown to stationary phase, and cells were transferred into 200 ml bottles and heat-killed in a water bath at 70 °C for at least 1 h. Cells were transferred into 50 ml sterile Falcon tubes and concentrated by centrifugation using an Allegra 25 R centrifuge (Beckman Coulter, Inc., Brea, CA, USA) equipped with a TS-5.1-500 swinging bucket rotor (Beckman Coulter) at 5000 r.p.m for 15 min. Supernatant was discarded and cells re-suspended using sterile Milli-Q water by vortexing. Cells were centrifuged and washed at least three times. After the final wash, cells were re-suspended in sterile Milli-Q water, and aliquots were stored frozen in 2 ml Eppendorf tubes until use. The effectiveness of the heat-killing process was verified by inoculating aliquots of the HKB into rich nutrient broth and monitoring for growth.
In order to label bacteria with stable isotopes, the glucose and ammonium in the M9 medium were substituted with d-glucose-1-13C (glucose 4% final concentration, 99 atom % 13C, Sigma-Aldrich Corp., St Louis, MO, USA) and ammonium-15N chloride (18 mm final concentration, 98 atom % 15N, Sigma-Aaldrich Corp., St Louis, MO, USA). Growth, heat-killing and processing of the labeled bacteria followed the same protocol described above.
Ochromonas cultures and experimental incubations
Cultures of Ochromonas sp. strain BG-1 were maintained axenic and bacteria-free on a modified DY-V recipe: medium was prepared following the Provasoli-Guillard National Center for Marine Algae and Microbiota protocol (see https://ncma.bigelow.org/algal-recipes) but without either MES buffer or nitrate, and with the addition of sodium bicarbonate at 95 μM; the sole inorganic source of nitrogen in this medium was ammonium chloride at 50 μM. Prior to the experimental incubations, an axenic Ochromonas culture was supplemented with HKB in order to increase the abundance of Ochromonas. The resulting culture was used as the inoculum for the experimental incubations once HKB had been depleted for 2–3 days (4–5 days of growth following inoculation). Three experiments combining different sources of labeled substrates/prey and light conditions were performed:
Experiment 1: Ochromonas was grown in triplicate cultures with labeled-inorganic substrates; ammonium-15N chloride (98 atom % 15N, Sigma-Aldrich Corp.) was added as 50% of the total ammonium (25 μM ammonium-15N and 25 μm ammonium-14N) and sodium bicarbonate-13C (98 atom % 13C, Sigma-Aldrich Corp.) was added as 100% of the total bicarbonate (95 μm). A separate set of triplicate control cultures with unlabeled ammonium and bicarbonate was run in parallel. Initial abundances of Ochromonas in these cultures were ~5 × 103 cells ml−1. Unlabeled HKB were added at abundances of 5 × 107 HKB ml−1 to both sets of triplicate cultures at the time of inoculum with the alga to ensure that prey was initially present at high abundance. Cultures were incubated at 20 °C with constant illumination and constant mixing using a magnetic stirrer to avoid settlement of non-motile HKB. We chose stirring based on preliminary work with other Ochromonas strains in the lab that compared constant gentle stirring vs constant gentle shaking. In these preliminary experiments, no significant differences were observed in growth rates between these methods of mixing.
Experiment 2: Ochromonas was grown in triplicate cultures with 13C- and 15N-labeled HKB. A separate set of triplicate control cultures with unlabeled HKB was run in parallel. Unlabeled ammonium and bicarbonate were added to the media at the same concentrations as in Experiment 1, and initial abundances of the alga (~ 5 × 103 cells ml−1) and HKB (~ 5 × 107 HKB ml−1) were also the same. Cultures were incubated at 20 °C with constant illumination and constant mixing using a magnetic stirrer.
Experiment 3: Ochromonas was grown in triplicate cultures with 13C- and 15N-labeled inorganic substrates at the same concentrations previously used, unlabeled HKB at the same starting abundances, and culture conditions exactly as in Experiment 1, except that the incubation was carried out in continuous darkness. A set of control cultures with unlabeled ammonium and bicarbonate was incubated in parallel in continuous darkness.
Cultures were sampled daily for determinations of HKB and Ochromonas abundance and chlorophyll a (Chl a) concentration. Samples for counts of HKB were preserved with formaldehyde (3.7% final concentration) and stored at 4 °C prior to staining with DAPI using standard methods (Porter and Feig, 1980). HKB were enumerated using an Olympus BX-51 epifluorescence microscope (Olympus Scientific Solutions Americas, Inc., Waltham, MA, USA). Algal densities were determined from 1 ml samples preserved with 50 μl of Lugol’s solution and stored at 4 °C until enumeration by light microscopy using a Palmer-Maloney chamber. Chl a was measured from 50 ml samples filtered onto 25 mm diameter glass fiber filters (GF/F, Whatman), extracted with 4 ml 100% acetone at −20 °C overnight in the dark, and measured using a non-acidification protocol with a Trilogy Lab fluorometer (Turner Designs, Sunnyvale, CA, USA; Welschmeyer, 1994).
Samples for nutrients (ammonium and soluble reactive phosphate) were collected at the time of inoculation and periodically throughout the course of the experiments. Approximately 15 ml of sample was filtered through a 0.2 μm filter into a clean glass vial that had been previously triple-rinsed with 0.2 μm filtered Milli-Q water then triple-rinsed with 0.2 μm filtered sample water. Duplicate blanks of 0.2 μm filtered Milli-Q water were stored and analyzed for each sampling date. The vials were stored frozen until analysis at the Marine Science Institute Analytical Lab at the University of California Santa Barbara (MSI-UCSB) by flow injection analysis using a QuikChem 8000 (Lachat Instruments Div., Zellweger Analytics, Inc., Hach Company, Loveland, CO, USA).
Bulk particulate carbon and nitrogen, and 13C- and 15N-isotopes, were determined from 30–50 ml of the cultures filtered onto pre-combusted glass fiber filters (GF/F, Whatman). Samples were collected at the time of inoculation and when HKB abundance had been reduced three orders of magnitude. Carbon and nitrogen were also measured in 50–150 μl aliquots of the stock suspensions of HKB. Blank filters consisting of 20 ml of 0.2 μm filtered Milli-Q water were also analyzed. Filters were dried at 60 °C. Analysis of CHN was done at the MSI-UCSB using a CEC 440HA (Exeter Analytical; EA) following standard protocols, and an acidification step with HCl to remove inorganic carbon. The C- and N-isotopic composition of the particulate organic matter was determined as CO2 and N2 released by flash combustion using a Finnigan DELTAplus Advantage in Continuous Flow Mode coupled to an isotope ratio mass spectrometer (IRMS) Costech ECS 4010 Elemental Combustion System.
NanoSIMS analysis
NanoSIMS analysis was conducted in order to examine the source of carbon and nitrogen (that is, inorganic vs HKB) incorporated by Ochromonas. Sample preparation for nanoSIMS included 2 ml of culture collected at the time of inoculation and at 48 h (the end of rapid growth of Ochromonas when bacterial prey had been exhausted). Samples of HKB stocks were also collected. Samples were preserved with 2X EM grade glutaraldehyde (2.5% final concentration) and stored at 4 °C until analysis. Cells were deposited onto silicon wafers (7 × 7 mm, 725 +/−25 μm thick, P-type/Boron 0.028 Ω-cm resistivity; Active Business Company GmbH, Munich Germany), washed with Milli-Q water and air dried. NanoSIMS was performed at the Caltech Microanalysis Center using a Cameca NanoSIMS 50 L instrument. Individual cells were located using the nanoSIMS CCD camera and random areas on the wafer were chosen for analysis. Algal cells were pre-sputtered with a 30 pA primary Cs+ beam current for 1 frame (~12 min) and then analyzed using a 1 pA primary Cs+ beam current with a nominal spot size of 100 nm.
For all analyses, the beam was rastered over a square region of 10 × 10 μm for 12 min per analytical frame. Pre-sputtering was carried out on a larger region of at least 13 × 13 μm to make sure that the analytical frame was fully within the pre-sputtered area. Four masses, 12C2−, 12C13C−, 12C14N− and 12C15N−, were collected in parallel using electron multipliers. Secondary electrons were also collected during the analysis, providing secondary electron images of the cells. Ion images were recorded at 512 × 512 pixel resolution with a dwell time of 2.5 ms per pixel and at least 4 frames per analysis, which were merged into a single frame during post-processing. A total of 33 Ochromonas cells from the end of the rapid growth phase of the alga were analyzed, with a minimum of 4 cells analyzed per treatment. Unlabeled HKB with a known 15N and 13C composition were measured at the start and end of all nanoSIMS measurements in order to determine the magnitude of instrumental isotope fractionation and drift during the analysis. The average nanoSIMS acquired 15N/14N and 13C/12C ratios were then compared and corrected with the elemental analysis-IRMS acquired values for the unlabeled HKB. The offset between the nanoSIMS and the elemental analysis-IRMS resulted in a correction factor of 1.1165 for 13C ratios and 1.0724 for 15N ratios. 13C and 15N cellular enrichments measured by bulk elemental analysis-IRMS and single-cell nanoSIMS analyses for the HKB were: 15N FHKB was 0.779±0.011 (bulk elemental analysis-IRMS) or 0.901±0.001 (nanoSIMS) and 13C FHKB was 0.041±0.00003 (bulk elemental analysis-IRMS) or 0.050±0.001 (nanoSIMS). Error values indicate standard errors.
The raw data from ion images were processed using the open-source MATLAB plugin Look@NanoSIMS (Polerecky et al., 2012). Ion images from multiple frames were corrected for dead time and quasi-simultaneous arrival (QSA) effect, accumulated and aligned. Each cell was defined as a region of interest (ROI) using the 14N12C− image and compared with the corresponding secondary electron image as a guide.
Calculations
The growth rates of Ochromonas were calculated between sampling intervals using the formula:

where μ is growth rate, (Ochromonasf) is the final abundance of Ochromonas, (Ochromonasi) is the initial abundance of Ochromonas and Tf and Ti are the final and initial sampling times in hours, respectively. Ingestion rates were calculated for the alga up to time when bacterial abundances were greatly reduced as in (Sanders et al., 2001), using the formula:

where I is ingestion rate, Δ (Ochromonas) the change in abundance of the alga, and Tf and Ti are the final and initial sampling times in hours, respectively.
Molar concentrations of nitrogen in HKB were calculated from CHN values of known abundances of HKB. Molar concentrations of nitrogen in Ochromonas were calculated from CHN values of known abundances of the alga in the inoculum at time 0, and at successive time points. Molar concentrations of dissolved inorganic nitrogen were obtained from ammonia concentrations in the cultures. Nitrogen molar concentrations associated with both HKB and Ochromonas biomass at the time of inoculum and when HKB were grazed to very low abundances (after 48 h, at which time nitrogen associated with HKB was negligible), were used to calculate grazing assimilation efficiency.
For nanoSIMS, all ion ratios were converted to fractional abundance using the relationship Fsample=Rsample/1+Rsample, where F is the fractional abundance and R is the ratio of either 13C/12C or 15N/14N. The carbon isotope data were collected using the molecular ions 13C12C- and 12C2- and to accurately calculate the 13C fractional abundance from our raw ion counts, the Rsample was multiplied by 0.5.
Finally, to determine the sources responsible for the isotopic enrichment in Ochromonas cells (fHKB or fInorg.), the following isotope mass balance was used for both carbon and nitrogen calculations:

assuming that:

where FOchromonas is the 13C or 15N fractional abundance for the alga, FHKB is the 13C or 15N fractional abundance for the heat-killed bacteria, and FInorg is the 13C or 15N fractional abundance of the inorganic substrate. Nitrogen FInorg was assumed to be either 0.49 for treatments with 15N-ammonium chloride (based on molar additions), or 0.00367 for non-labeled treatments (based on natural abundance); Carbon FInorg was assumed to be 0.98 for treatments with 13C-bicarbonate or 0.0109 for non-labeled treatments. Using equations (iii) and (iv) we solved for fHKB (the fraction of HKB) and fInorg (the fraction of inorganic nitrogen or carbon) in the different experimental treatments.
Results
Growth of Ochromonas sp. strain BG-1
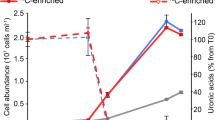
Initial abundances of Ochromonas in all experimental incubations ranged from 5 × 103 to 1.2 × 104 Ochromonas cells ml−1. Rapid growth of Ochromonas lasted ~2 days (Figure 1a). The growth rate throughout this entire growth phase ranged from 1.7 to 2.0 day−1 (Table 1), and cell abundances increased 30- to 60-fold (Figure 1a). HKB abundances decreased 3 orders of magnitude during this period of rapid algal growth, from the average inoculum of 5 × 107 HKB ml−1 to an average value of 9 × 104 HKB ml−1 after 48 h of algal grazing (Figure 1b). HKB abundances were too low to be counted confidently after 100 h of incubation. Although algal abundance increased many-fold in the first 48 h, total Chl a increased only 1.4- to 3-fold for algae incubated in the light (Figure 1c), effectively resulting in decreases in the chlorophyll concentration alga−1 in all treatments (Figure 1d). Ammonium and phosphate during this period showed no net removal, but rather accumulated substantially in the medium during the first 48 h of the experiments (Figure 2). Blank measurements of 0.2-μm filtered Milli-Q water were consistently two orders of magnitude lower than culture samples, or below the detection limit.
Cell abundances of Ochromonas sp. strain BG-1 (a), abundances of heat-killed bacteria, HKB (b), Chl a concentration in the cultures (c) and Chl a alga−1 (d) in three experiments. Average and standard error values for three replicates are represented. Solid lines represent cultures incubated in 24 h light regimes, dotted lines indicate cultures in continuous darkness. Circles represent cultures incubated with stable isotope labeled HKB and non-labeled inorganics, triangles indicate cultures with stable isotope labeled inorganics and non-labeled HKB. Squares are control cultures grown with unlabeled inorganics and unlabeled HKB.
Ammonium and phosphate concentrations (μM) in three experiments with Ochromonas grown with heat-killed bacteria (HKB) and inorganic nutrients. Averages and standard errors for three replicates are represented. Solid lines represent cultures incubated in 24 h light regimes, dotted lines cultures in continuous darkness. Circles represent cultures incubated with stable isotope labeled HKB and non-labeled inorganics, triangles indicate cultures with stable isotope labeled inorganics and non-labeled HKB, and squares are control cultures grown with both unlabeled inorganics and unlabeled HKB.
Ochromonas abundances in cultures incubated in the light did not increase significantly beyond the first 48 h of incubation (Figure 1a). Nevertheless, total Chl a concentrations continued to increase in those cultures after 48 h, resulting in increases in chlorophyll content cell−1 (Figures 1c and d). Concurrently, ammonium concentrations decreased in cultures receiving light following 48 h of rapid algal growth, although phosphate concentrations continued to increase (Figure 2). Cultures incubated in continuous darkness exhibited low total Chl a concentrations throughout the entire experiment, as well as very low cellular Chl content (dotted lines in Figures 1c and d). Both ammonium and phosphate concentrations continued to increase throughout the experiment in Ochromonas cultures incubated in continuous darkness (Figure 2).
Consumption of HKB by Ochromonas averaged 15 bacteria cell−1 h−1 through the 2 days of rapid growth of the alga (Table 1). Ingestion rates were higher for the first 24 h, with values up to 50 bacteria cell−1 h−1. Ochromonas reduced HKB abundances three orders of magnitude after 2 days, to less than 105 HKB ml−1. A grazing efficiency for nitrogen by the alga was estimated from algal and bacterial abundance measurements and the molar concentrations of nitrogen contained in HKB and Ochromonas. On the basis of those estimates, ~50% of nitrogen contained in HKB was assimilated by Ochromonas when grown in the light; cultures that were grown in the dark presented a slightly lower nitrogen transfer efficiency, with up to 30% of nitrogen in HKB assimilated by Ochromonas (Figure 3).
Nitrogen mass balance for the three different experiments with Ochromonas. All nitrogen at the beginning of the experiments was either present as organic nitrogen (associated with Ochromonas or HKB) or inorganic nitrogen in the medium (ammonium). Nitrogen associated with Ochromonas or the inoculum of HKB was calculated from CHN analysis of the two populations and cell abundances in the cultures, with values transformed to μM. Dissolved ammonium was measured and transformed to μM concentrations. After ~48 h of incubation, virtually all HKB were consumed by Ochromonas, and the nitrogen contained within HKB was assimilated into cells (area of column with dots at time 48 h) or released into the medium. Arrows represent an estimate of the percentage of nitrogen associated with HKB at time 0 and assimilated by Ochromonas at time 48 h, assuming that the excess nitrogen was released as ammonium.
Stable isotope analysis
Incubations were carried out with either labeled inorganic nitrogen and carbon (as 15N-ammonium chloride and 13C-bicarbonate) or labeled heat-killed bacteria (HKB; previously grown on labeled substrates; 15N-ammonium and 13C-glucose) in order to determine the sources of nitrogen and carbon incorporated by Ochromonas. Isotopic ratios measured by nanoSIMS were consistent both within single cells and between cells within the same treatment. The error associated with repeated measures of the same cell (measured as a Poisson error) was always at least 2 orders of magnitude lower than the measured isotopic fractionation. For cells within the same treatment, the standard error was at least 1 order of magnitude lower than their average isotopic ratio. At the end of the period of rapid growth, all three experiments presented a significantly higher fractional abundance of both 15N and 13C in replicates incubated with an enriched isotopic substrate compared to the control replicates (ANOVA, for each treatment and isotope P<0.01). Ochromonas was most enriched in the incubations with labeled HKB (fractional abundance for 15N=0.793±0.007, 13C=0.044±0.001; Figures 4a and b) followed by the incubations amended with 15N-ammonium and 13C-bicarbonate in the light (fractional abundance for 15N=0.028±0.003, 13C=0.017±0.001; Figures 4a and b). The experiment performed with labeled inorganic substrates in the dark yielded low values (fractional abundance for 15N=0.012±0.0004, 13C=0.012±0.0001; Figures 4a and b).
(a) Examples of cells analyzed by nanoSIMS, showing both unlabeled and labeled Ochromonas. The first row shows images of the alga obtained through secondary ion emission, the second row shows the fractional abundance for 13C, and the third row shows images of the fractional abundance for 15N. Note the different panel scales for 13C and 15N fractional abundances: the 13C fractional abundance color scale ranges from 0.02 to 0.1; the 15N color scale range varies among the four panels as differences were too wide to represent with a single scale for all panels. Columns in (a), from left to right, show cells in the control treatment (no stable isotope labeling), cells grown in the presence of stable isotope labeled inorganic carbon and nitrogen with unlabeled HKB (Experiment 1), cells grown in the presence of stable isotope labeled HKB and unlabeled inorganic carbon and nitrogen (Experiment 2), and cells grown in the presence of stable isotope labeled inorganic carbon and nitrogen with unlabeled HKB but incubated in continuous darkness (Experiment 3). (b) Plot of average (± SE) fractional abundances for 15N and 13C measured by nanoSIMS in cells from inoculum HKB, inoculum Ochromonas and Ochromonas at the end of rapid growth (>48 h). Values for the three experiments are presented, including the stable isotope labeled and non-labeled replicates; note that error bars are often smaller than the data points. (c) and (d) show the fractional abundances for 15N and 13C, respectively, measured by nanoSIMS vs the fractional abundances measured by bulk IRMS. Samples for nanoSIMS and IRMS were collected at the same time. Diagonal lines in the plots correspond to a 1:1 equivalence.
There was good agreement between nanoSIMS and bulk elemental analysis-IRMS measurements showing the same trends in enrichment (Figures 4c and d), although there was a somewhat greater offset between these measurements for samples that were highly enriched, particularly for 13C carbon. Additional bulk elemental analysis-IRMS measurements were collected after 48 h, at time points beyond the period of rapid growth of the alga when HKB had been reduced to insignificant abundances. These measurements revealed a decrease in F for 13C in the treatments with labeled HKB (Experiment 2 in Figure 5), while the treatments with labeled inorganic nitrogen and carbon in the light exhibited a slight increase in fractional 15N (Experiment 1 in Figure 5). Fractional abundances for the culture incubated in continuous darkness did not show significant changes after the depletion of HKB (Experiment 3 in Figure 5).
Plots of average (±s.e.) fractional abundances for 15N and 13C measured by bulk IRMS for inoculum HKB, inoculum Ochromonas, and Ochromonas at the end of rapid growth (~ 48 h) plus two additional values for Ochromonas after the period of rapid growth (>48 h). Numbers above symbols indicate the sampling time (h); non-labeled time stamps have been removed for ease of visualization because they presented similar values around the natural isotopic abundance of 15N and 13C (red symbols). Values for the three experiments are presented, including the labeled and non-labeled replicates. Note that axis scales among the three panels differ.
We used the stable isotope mass balance calculations (iii) and (iv) described in the Material and Methods in order to determine the relative contributions of either organic or inorganic nitrogen and carbon sources during mixotrophic growth of Ochromonas. The nanoSIMS measurements of 15N and 13C enrichment of algal cells recovered from the different incubation conditions revealed that most of the nitrogen and carbon assimilated by Ochromonas was derived through bacterivory of the HKB, rather than CO2 fixation and ammonium assimilation. The results of Experiment 1 (incubated in light) and Experiment 3 (incubated in continuous darkness) using heavy isotope-labeled inorganics and unlabeled HKB indicated that 95 or 98% of algal nitrogen, and 99.4 or 99.9% of algal carbon, respectively, was derived from HKB. Experiment 2, using heavy isotope-enriched bacteria and incubated in the light, yielded slightly lower values of incorporation of nitrogen (88%) and carbon (84%) from HKB, but bacteria were still the major source of nitrogen and carbon in all experiments.
Discussion
Growth and nutrient dynamics of Ochromonas sp., strain BG-1
Phagotrophy by phototrophic flagellates is believed to confer significant ecological advantages on algae that exhibit the behavior (Flynn and Mitra, 2009), but there also appear to be limitations (although largely uncharacterized) for conducting mixed nutrition simultaneously in a single cell (Raven, 1997). These constraints may involve costs or tradeoffs for maintaining dual cellular machineries (compared to specialized competitors), or perhaps cross-talk between anabolic and catabolic biochemical pathways that confound the performance of both activities simultaneously. Unfortunately, there is virtually no quantitative information on the costs and benefits of mixotrophic behaviour, nor whether both processes are carried out sequentially or simultaneously by these species. Temporal partitioning might be a mechanism for maintaining both abilities in minute cells (albeit putting one process ‘on hold’ at any given time). Assessing whether both behaviors are occurring simultaneously, the specific physiological benefit(s) for heterotrophic nutrition in mixotrophic algae, and the responsiveness of this behavior to abiotic and biotic variables are aspects of mixotrophic nutrition that have been difficult to establish using traditional approaches and methods.
We studied the mixotrophic chrysophyte Ochromonas sp. strain BG-1 because it can be grown in axenic culture (bacteria-free) and it has been previously reported to be a predominantly heterotrophic organism that attains high growth rates only in the presence of bacteria (Sanders et al., 2001). Growth in axenic culture prevented potentially complicating biological interactions and elemental flows resulting from the activities of other living microorganisms in the cultures, thereby enabling a comparison between nanoSIMS and bulk IRMS measurements and a better understanding of the specific sources of carbon and nitrogen used for growth by the alga.
Additionally, the use of a single inorganic source of nitrogen simplified our experimental design so that the only sources of nitrogen in the medium were ammonium or HKB. Algae generally possess mechanisms for the uptake and assimilation of both ammonium and nitrate, but transcriptomic data indicate that some chrysophytes, including Ochromonas sp. strain BG-1, might be missing the genetic capability for nitrate assimilation (Terrado et al., 2015; Lie et al., 2017). Furthermore, MES buffer was not added to the medium in our experiments because this might have provided an alternative source of nitrogen. MES might also have constituted a source of organic carbon for the alga (Sanders et al., 2001), and its elimination ensured that the only sources of carbon in the medium were either bicarbonate or HKB. Transcriptomic analysis from experiments carried out in the same way as the ones presented in this manuscript show that the photosynthetic machinery of Ochromonas sp. strain BG-1 is expressed and upregulated in the presence of light (Lie et al., 2017). This simplified approach, combined with stable isotope labeling, allowed us to determine which source(s) of nitrogen and carbon were used for growth by the alga.
Significant growth rates of Ochromonas in the present study were achieved only while HKB were abundant as prey (Figure 1a). Chl a dynamics in the culture also reflected the high growth rates due to grazing on HKB, as the concentration of Chl a cell−1 decreased by one order of magnitude during the first 48 h of incubation for cultures grown in the light as well as in continuous darkness (Figure 1d). These changes in cellular chlorophyll might be related to a dilution of Chl a inside the cells due to the high growth rates of the alga (Hansen et al., 2000), so that the reduction of Chl a cell−1 was a consequence of fast growing rates of the alga and not a direct reduction of chlorophyll biosynthesis rate. Nevertheless, it is also possible that there was some regulation of chlorophyll biosynthesis when HKB were present as transcriptomic analysis on this alga suggests upregulation of genes related to chlorophyll synthesis in the presence of light when HKB were depleted (Lie et al., 2017). In any case, these observations are in agreement with previous studies of Ochromonas (Pringsheim, 1952; Sanders et al., 2001) that observed a well-developed heterotrophic capability, suggesting that carbon fixation and perhaps other cellular structures and processes involved in photosynthesis are reduced when growing mixotrophically, as observed for some other algae (Wan et al., 2011).
Dissolved ammonium, as well as phosphate, accumulated in the Ochromonas cultures during the first 48 h of the experiments, when HKB were actively grazed (Figure 2). This result indicates that excess nitrogen and phosphorous from grazed HKB were excreted by the alga. Mass balance calculations based on changes in prey/algal abundances and their cellular nitrogen contents indicated that up to 50% of the nitrogen contained in consumed HKB was assimilated by the alga, while a considerable amount of the excess nitrogen was released primarily as ammonium during the period of active bacterial grazing (Figure 2). These values for nitrogen assimilation (and excretion) are consistent with assimilation efficiencies of heterotrophic protists of similar size (Taylor, 1982; Caron and Goldman, 1990), consistent with the conclusion that Ochromonas was growing predominantly as a heterotroph when prey were abundant. Further, transcriptomic analyses have demonstrated that different ammonium transporters are expressed by Ochromonas sp. strain BG-1 growing on HKB compared to growth after bacteria have been grazed to very low abundances (Lie et al., 2017). It therefore appears that transporters for the export of ammonium out of the cell may be different than those used for ammonium uptake, as has been observed for other organisms (Shnaiderman et al., 2013).
Interestingly, concentrations of ammonium, but not phosphate, decreased in the medium once the HKB had been removed by grazing (that is, after 48 h; Figure 2) when Ochromonas was grown in the light. This result appears to indicate that the alga actively took up ammonium (but not phosphate) from the medium when bacteria were no longer available and photosynthesis was induced (Figure 1d). In contrast, both ammonium and phosphate in the cultures grown in the dark continued to rise throughout the experiment (dotted lines in Figure 2). No significant net algal population growth occurred following depletion of prey even in the light, and the explanation for the dichotomy in the uptake of these two elements is unclear. We speculate that ammonium was taken up because it was specifically required for rebuilding the photosynthetic machinery of the cell.
The continued appearance of phosphate in the culture medium during the later part of the experiments contrasts with studies of some other Ochromonas species that have reported an uptake of phosphate when the alga is growing autotrophically (Rothhaupt, 1996). The lack of phosphate uptake by strain BG-1 might indicate that this Ochromonas was incapable of effective phosphate uptake (which might also explain, in part, the poor phototrophic growth capacity of this strain), or that phosphorus was not needed in significant amounts for cellular reorganization associated with the change to phototrophic growth, and therefore uptake was not stimulated by the change to phototrophy. It is unlikely that the continued increases in the concentration of phosphate were due to the decomposition of dissolved organic phosphorus compounds in the medium because the cultures had no live bacteria.
Inferences from stable isotope probing experiments
Stable isotope analysis (nanoSIMS and bulk elemental analysis-IRMS) revealed that both inorganic (13C-bicarbonate and 15N-ammonium) substrates and 13C/15N-labeled HKB were assimilated by Ochromonas, contributing to 15N and 13C cellular enrichment after 48 h of incubation (Figure 4). However, the magnitude of enrichment from inorganic substrates or HKB indicated that, during mixotrophic growth, the primary sources of carbon and nitrogen were derived from phagotrophy. Isotopic mass balance indicated that 88–95% of the nitrogen and 84–99% of the carbon were derived from HKB when Ochromonas was growing mixotrophically in the light. An enrichment of 13C was observed in algae grown in the light compared to the dark (Experiment 1 vs Experiment 3) when 13C-bicarbonate was available (Figure 4). However, the calculated contribution of photosynthetic carbon fixation was only 1–10% of the carbon assimilated into biomass. The greater efficiency of nitrogen incorporation from prey observed in the light relative to continuous darkness (Figure 3; Experiments 1, 2 vs Experiment 3) suggests that light played a role, albeit a minor one, in phagotrophic efficiency of the alga. Therefore, despite putative reductions in the photosynthetic machinery of Ochromonas growing phagotrophically on HKB (as evidenced by low cell quotas of Chl a; Figure 1d), light did have a minor and positive impact on algal nutrition. Because the amount of carbon fixed by photosynthesis represented a small fraction of carbon assimilated by the alga when growing on HKB, we speculate that the photosynthetic apparatus might be providing energy rather than carbon for cellular material, as it has been hypothesized for Ochromonas danica (Wilken et al., 2014).
Our isotopic mass balance calculations have two caveats. First, we did not control the pH evolution within the cultures, which would have given better insights on the carbonate equilibrium that might have been affected by respiration and carbon fixation, and exchange with the atmosphere. As such, our estimate based on the experiments using labeled inorganic carbon might have underestimated the amount of inorganic carbon fixed by Ochromonas sp. BG-1 (1% according to the isotopic mass balance). In any case, the experiment using labeled HKB should not have been affected by this caveat, and the estimate of 10% of carbon derived from inorganic substrate is likely realistic. Second, Ochromonas is considered an inefficient carbon fixer due to missing carbon concentration mechanisms (Maberly et al. 2009) that increase CO2 concentration via the transport of CO2 and/or bicarbonate to the RubisCO enzyme (Raven et al., 2008). On the other hand, transcriptomic analysis have shown that the photosynthetic machinery of strain BG-1 is functional (Lie et al., 2017), and our experiments using labelled bicarbonate showed a significant enrichment of the 13C fractional abundance in Ochromonas (Figures 4 and 5); therefore, Ochromonas sp. strain BG-1 has some capability to use inorganic carbon, albeit inefficiently.
Nevertheless, the strong heterotrophic activity of Ochromonas while growing mixotrophically is likely to increase the intracellular CO2 pool as well as its flux. As a rule of thumb, it is considered that heterotrophic protists assimilate 40% of ingested organic matter, while they release 30% and respire another 30% (Sleigh, 1989). Based on this, the total amount of carbon released by Ochromonas as CO2 during exponential growth could be as high as the total bicarbonate added at the beginning of the incubation, which has consequences for the isotopic mass balance we have presented. If we assume that the isotopically enriched CO2 derived from the HKB respiration is available at the same levels as the dissolved inorganic carbon, the incubation performed with Ochromonas and labeled HKB indicated that ~84% of carbon was derived from the HKB. Up to 20% of the assimilated carbon derived from HKB could actually correspond to carbon that was initially respired and then fixed by Ochromonas. If correct, respiration derived from the phagotrophic activity would act as a carbon concentration mechanism of sorts for this Ochromonas. While the primary source of carbon for Ochromonas growing mixotrophically was derived from HKB, a non-negligible amount might have been derived from the respiration and then CO2 fixation of the bacterial biomass.
Ochromonas shifted its metabolism towards autotrophy when incubated in the light, but only after it had depleted the HKB in the cultures (≈ 48 h of growth). This shift was reflected in the bulk IRMS 13C fractional abundance for the treatment using labeled HKB where a reduction was observed between 48 h and 145 h (Experiment 2 in Figure 5), indicative of the incorporation of unlabeled carbon into algal biomass via light processes. Chrysophytes are generally considered poor autotrophic carbon fixers due to poor carbon concentration mechanisms (Maberly et al., 2009). Nonetheless, these results indicate that there was a significant level of inorganic carbon assimilation. A comparison of cultures grown in the light (Experiment 1 in Figure 5) and continuous darkness (Experiment 3 in Figure 5) revealed that the 15N fractional abundance of the dark cultures did not change after 95 h while cultures in the light continued to enrich in 15N, indicating that Ochromonas continued to assimilate nitrogen to maintain its metabolism once HKB were depleted. These results were consistent with observed decreases in the concentration of ammonium in the medium during this time period (Figure 3a), although the continued appearance of phosphate in the medium during this period is unexplained.
We obtained good overall agreement between the bulk isotope measurements and nanoSIMS measurements for nitrogen, consistent with observations in previous studies (Popa et al., 2007; Orphan and House, 2009; Kopf et al., 2015; Figure 4c). However, the bulk measurements were somewhat lower in terms of fractional abundance values for carbon, particularly for highly enriched samples (Figure 4d). We speculate that the differences between nanoSIMS and the bulk isotope measurements for carbon may be related to the fact that nanoSIMS samples were preserved with glutaraldehyde, whereas the samples for bulk analysis were not. Fixation has been shown to influence cellular carbon (Musat et al., 2014), although we might have expected this to dilute the 13C in the nanoSIMS measurements relative to the bulk values. A more likely explanation is that the single-cell measurements are not influenced by cellular debris in the culture that may be less enriched. The bulk 13C value may be diluted by these components relative to the nanoSIMS measurements, implying that the nanoSIMS data may more accurately reflect carbon and nitrogen uptake by the algae. However, cell to cell variability may also have contributed to the minor differences between the bulk and nanoSIMS measurements.
The use of nanoSIMS in this study represents its first application towards the study of carbon and nutrient fluxes in a mixotrophic alga, and has enabled a better understanding of carbon and energy acquisition by this species, and cellular metabolism. Our findings expand information available from traditional analyses of Ochromonas sp. strain BG-1 grown under various conditions of light and prey availability (Sanders et al., 2001), confirming that most of the nitrogen and carbon used for growth are obtained through its bacterial prey. Although the results cannot be directly extrapolated to all species along the continuum of algae with different mixotrophic strategies, our work validates the use of stable isotope probing experiments and nanoSIMS to better understand the metabolic underpinnings of mixotrophy in one species. Moreover, it provides an approach for assessing mixotrophic nutrition in environmental samples. Ochromonas sp. strain BG-1 provided an ideal model system for comparing bulk isotope analysis to nanoSIMS because bacteria were quickly removed by grazing within the first 48 h of the experiments. The agreement between these two measurements demonstrates that nanoSIMS accurately captured the dynamics of carbon and nutrient acquisition in this mixotroph, and could therefore be more broadly applied to explore mixotrophy in complex mixed communities where bulk measures would be insufficient to capture these dynamics. This and future detailed studies will continue to yield improvements in our understanding of the nutrition of mixotrophic algae.
References
Andersen KH, Aksnes DL, Berge T, Fiksen Ø, Visser A . (2015). Modelling emergent trophic strategies in plankton. J Plankton Res 37: 862–868.
Andersen RA. (2007). Molecular systematics of the Chrysophyceae and Synurophyceae. In: Brodie J, Lewis J (eds). Unravelling the Algae: the Past, Present, and Future Of Algal Systematics. CRC Press: Boca Rotan, FL, USA, pp 285–313..
Andersson A, Falk S, Samuelsson G, Hagström Å . (1989). Nutritional characteristics of a mixotrophic nanoflagellate, Ochromonas sp. Microb Ecol 17: 117–128.
Azam F, Malfatti F . (2007). Microbial structuring of marine ecosystems. Nat Rev Microbiol 5: 782–791.
Boraas ME, Seale DB, Holen D . (1992). Predatory behavior of Ochromonas analyzed with video microscopy. Arch Hydrobiol Beih 123: 459–468.
Boxer SG, Kraft ML, Weber PK . (2009). Advances in imaging secondary ion mass spectrometry for biological samples. Ann Rev Biophys 38: 53–74.
Caron DA, Goldman JC. (1990). Protozoan nutrient regeneration. In: Capriulo GM (ed). Ecology of Marine Protozoa. Oxford University Press: New York, NY, USA, pp 283–306..
Caron DA, Porter KG, Sanders RW . (1990). Carbon, nitrogen and phosphorus budgets for the mixotrophic phytoflagellate Poterioochromonas malhamensis (Chrysophyseae) during bacterial ingestion. Limnol Oceanogr 35: 433–443.
Caron DA, Sanders RW, Lim EL, Marrasé C, Amaral LA, Whitney S et al. (1993). Light-dependent phagotrophy in the freshwater mixotrophic chrysophyte Dinobryon cylindricum. Microb Ecol 25: 93–111.
Carpenter KJ, Weber PK, Davisson ML, Pett-Ridge J, Haverty MI, Keeling PJ . (2013). Correlated SEM, FIB-SEM, TEM, and NanoSIMS imaging of microbes from the hindgut of a lower termite: methods for in situ functional and ecological studies of uncultivable microbes. Microsc Microanal 19: 1490–1501.
Chrzanowski TH, Foster BLL . (2014). Prey element stoichiometry controls ecological fitness of the flagellate Ochromonas danica. Aq Microb Ecol 71: 257–269.
del Campo J, Massana R . (2011). Emerging diversity within chrysophytes, choanoflagellates and bicosoecids based on molecular surveys. Protist 162: 435–448.
Finzi-Hart JA, Pett-Ridge J, Weber PK, Popa R, Fallon SJ, Gunderson T et al. (2009). Fixation and fate of C and N in the cyanobacterium Trichodesmium using nanometer-scale secondary ion mass spectrometry. Proc Natl Acad Sci USA 106: 6345–6350.
Flynn KJ, Mitra A . (2009). Building the 'perfect beast': modelling mixotrophic plankton. J Plankton Res 31: 965–992.
Flynn KJ, Stoecker DK, Mitra A, Raven JA, Glibert PM, Hansen PJ et al. (2013). Misuse of the phytoplankton-zooplankton dichotomy: the need to assign organisms as mixotrophs within plankton functional types. J Plank Res 35: 3–11.
Foster BLL, Chrzanowski TH . (2012). The mixotrophic protist Ochromonas danica is an indiscriminant predator whose fitness is influenced by prey type. Aq Microb Ecol 68: 1–11.
Gao D, Huang X, Tao Y . (2016). A critical review of NanoSIMS in analysis of microbial metabolic activities at single-cell level. Crit Rev Biotechnol 36: 884–890.
Hansen PJ, Skovgaard A, Glud RN, Stoecker DK . (2000). Physiology of the mixotrophic dinoflagellate Fragilidium subglobosum. II. Effects of time scale and prey concentration on photosynthetic performance. Mar Ecol Prog Ser 201: 137–146.
Hartmann M, Grob C, Tarran GA, Martin AP, Burkill PH, Scanlan DJ et al. (2012). Mixotrophic basis of Atlantic oligotrophic ecosystems. Proc Nat Acad Sci USA 109: 5756–5760.
Jones RI . (1994). Mixotrophy in planktonic protists as a spectrum of nutritional strategies. Mar Microb Food Webs 8: 87–96.
Jones RI . (2000). Mixotrophy in planktonic protists: an overview. Freshwater Biol 45: 219–226.
Kimura B, Ishida Y . (1989). Phospholipid as a growth factor of Uroglena americana, a red tide chrysophyceae in Lake Biwa. Nippon Suisan Gakk 55: 799–804.
Kopf SH, McGlynn SE, Green-Saxena A, Guan Y, Newman DK, Orphan VJ . (2015). Heavy water and 15 N labelling with NanoSIMS analysis reveals growth rate- dependent metabolic heterogeneity in chemostats. Environ Microbiol 17: 2542–2556.
Kopp C, Pernice M, Domart-Coulon I, Djediat C, Spangenberg JE, Alexander DTL et al. (2013). Highly dynamic cellular-level response of symbiotic coral to a sudden increase in environmental nitrogen. mBio 4: e00052–13.
Lie AAY, Liu Z, Terrado R, Tatters AO, Heidelberg KB, Caron DA . (2017). Effect of light and prey availability on gene expression of the mixotrophic chrysophyte, Ochromonas sp. BMC Genomics 18: 163.
Liu Z, Jones AC, Campbell V, Hambright KD, Heidelberg KB, Caron DA . (2015). Gene expression in the mixotrophic prymnesiophyte, Prymnesium parvum, responds to prey availability. Front Microbiol 6: 319.
Liu Z, Campbell V, Heidelberg KB, Caron DA . (2016). Gene expression demonstrates different nutritional strategies among three mixotrophic protists. FEMS Microbiol Ecol 92: fiw106.
Maberly SC, Ball LA, Raven JA, Sültemeyer D . (2009). Inorganic carbon acquisition by chrysophytes. J Phycol 45: 1052–1061.
Marley J, Lu M, Bracken C . (2001). A method for efficient isotopic labeling of recombinant proteins. J Biomol NMR 20: 71–75.
Mitra A, Flynn KJ, Tillmann U, Raven JA, Caron D, Stoecker DK et al. (2016). Defining planktonic protist functional groups on mechanisms for energy and nutrient acquisition: incorporation of diverse mixotrophic strategies. Protist 167: 106–120.
Morono Y, Terada T, Nishizawa M, Ito M, Hillion F, Takahata N et al. (2011). Carbon and nitrogen assimilation in deep subseafloor microbial cells. Proc Natl Acad Sci USA 108: 18295–18300.
Musat N, Stryhanyuk H, Bombach P, Adrian L, Audinot J-N, Richnow HH . (2014). The effect of FISH and CARD-FISH on the isotopic composition of 13C- and 15 N- labeled Pseudomonas putidacells measured by nanoSIMS. Syst Appl Microbiol 37: 267–276.
Orphan VJ . (2009). Methods for unveiling cryptic microbial partnerships in nature. Curr Opin Microbiol 12: 231–237.
Orphan VJ, House CH . (2009). Geobiological investigations using secondary ion mass spectrometry: microanalysis of extant and paleo-microbial processes. Geobiology 7: 360–372.
Polerecky L, Adam B, Milucka J, Musat N, Vagner T, Kuypers MMM . (2012). Look@NanoSIMS - a tool for the analysis of nanoSIMS data in environmental microbiology. Environ Microbiol 14: 1009–1023.
Popa R, Weber PK, Pett-Ridge J, Finzi JA, Fallon SJ, Hutcheon ID et al. (2007). Carbon and nitrogen fixation and metabolite exchange in and between individual cells of Anabaena oscillarioides. ISME J 1: 354–360.
Porter KG, Feig YS . (1980). The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr 25: 943–948.
Pringsheim EG . (1952). On the nutrition of Ochromonas. Q J Microsc Sci 3: 71–96.
Raven JA . (1997). Phagotrophy in phototrophs. Limnol Oceanogr 42: 198–205.
Raven JA, Cockell CS, De La Rocha CL . (2008). The evolution of inorganic carbon concentrating mechanisms in photosynthesis. Phil Trans R Soc B 363: 2641–2650.
Rothhaupt KO . (1996). Utilization of substitutable carbon and phosphorus sources by the mixotrophic chrysophyte Ochromonas sp. Ecology 77: 706–715.
Rottberger J, Gruber A, Boenigk J, Kroth PG . (2013). Influence of nutrients and light on autotrophic, mixotrophic and heterotrophic freshwater chrysophytes. Aquatic Microb Ecol 71: 179–191.
Sanders RW, Porter KG . (1988). Phagotrophic phytoflagellates. Adv Microb Ecol 10: 167–192.
Sanders RW, Caron DA, Davidson JM, Dennett MR, Moran DM . (2001). Nutrient acquisition and population growth of a mixotrophic alga in axenic and bacterized cultures. Microb Ecol 42: 513–523.
Sanders RW . (2011). Alternative nutritional strategies in protists: symposium introduction and a review of freshwater protists that combine photosynthesis and heterotrophy. J Euk Microbiol 58: 181–184.
Sarmento H . (2012). New paradigms in tropical limnology: the importance of the microbial food web. Hydrobiologia 686: 1–14.
Sheik AR, Brussaard CPD, Lavik G, Lam P, Musat N, Krupke A et al. (2014). Responses of the coastal bacterial community to viral infection of the algae Phaeocystis globosa. ISME J 8: 212–225.
Shnaiderman C, Miyara I, Kobiler I, Sherman A, Prusk D . (2013). Differential activation of ammonium transporters during the accumulation of ammonia by Colletotrichum gloeosporioides and Its effect on appressoria formation and pathogenicity. Mol Plant Microbe Interact 26: 345–355.
Sleigh MA . (1989) Protozoa and Other Protists. Cambridge University Press: Victoria, Australia.
Stoecker DK . (1998). Conceptual models of mixotrophy in planktonic protists and some ecological and evolutionary implications. Eur J Protistol 34: 281–290.
Taylor GT . (1982). The role of pelagic heterotrophic protozoa in nutrient cycling: a review. Ann Inst Oceanogr 58 (S): 227–241.
Terrado R, Monier A, Edgar R, Lovejoy C . (2015). Diversity of nitrogen assimilation pathways among microbial photosynthetic eukaryotes. J Phycol 51: 490–506.
Unrein F, Gasol JM, Not F, Forn I, Massana R . (2014). Mixotrophic haptophytes are key bacterial grazers in oligotrophic coastal waters. ISME J 8: 164–176.
Våge S, Castellani M, Giske J, Thingstad TF . (2013). Successful strategies in size structured mixotrophic food webs. Aq Ecol 47: 329–347.
Wan M, Liu P, Xia J, Rosenberg J, Oyler G, Betenbaugh M et al. (2011). The effect of mixotrophy on microalgal growth, lipid content, and expression levels of three pathway genes in Chlorella sorokiniana. Appl Microbiol Biotechnol 91: 835–844.
Ward BA, Follows MJ . (2016). Marine mixotrophy increases trophic transfer efficiency, mean organism size, and vertical carbon flux. Proc Natl Acad Sci USA 113: 2958–2963.
Welschmeyer NA . (1994). Fluorometric analysis of chlorophyll a in the presence of chlorophyll b and pheopigments. Limnol Oceanogr 39: 1985–1992.
Wilken S, Schuurmans JM, Matthijs HCP . (2014). Do mixotrophs grow as photoheterotrophs? Photophysiological acclimation of the chrysophyte Ochromonas danica after feeding. New Phytol 204: 882–889.
Worden AZ, Follows MJ, Giovannoni SJ, Wilken S, Zimmerman AE, Keeling PJ . (2015). Rethinking the marine carbon cycle: factoring in the multifarious lifestyles of microbes. Science 347: 1257594.
Acknowledgements
This collaborative research was supported by the Gordon and Betty Moore Foundation through Grant GBMF3299 to DAC and KBH at USC, and Grant (GBMF3780) to VJO at California Institute of Technology.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Terrado, R., Pasulka, A., Lie, AY. et al. Autotrophic and heterotrophic acquisition of carbon and nitrogen by a mixotrophic chrysophyte established through stable isotope analysis. ISME J 11, 2022–2034 (2017). https://doi.org/10.1038/ismej.2017.68
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2017.68
This article is cited by
-
Contribution of silica-scaled chrysophytes to ecosystems services: a review
Hydrobiologia (2023)
-
Broad phylogenetic and functional diversity among mixotrophic consumers of Prochlorococcus
The ISME Journal (2022)
-
Single-cell stable isotope probing in microbial ecology
ISME Communications (2022)
-
Microbial adaptation to different environmental conditions: molecular perspective of evolved genetic and cellular systems
Archives of Microbiology (2022)
-
Use of δ18O, δ 13C and NO3−to identify hydrogeochemical processes related to contamination in an aquifer located in central Mexico
Acta Geochimica (2022)