Abstract
Two novel chlorinated alkane-respiring Dehalobacter restrictus strains CF and DCA were isolated from the same enrichment culture, ACT-3, and characterized. The closed genomes of these highly similar sister strains were previously assembled from metagenomic sequence data and annotated. The isolation of the strains enabled experimental verification of predicted annotations, particularly focusing on irregularities or predicted gaps in central metabolic pathways and cofactor biosynthesis. Similar to D. restrictus strain PER-K23, strains CF and DCA require arginine, histidine and threonine for growth, although the corresponding biosynthesis pathways are predicted to be functional. Using strain CF to experimentally verify annotations, we determined that the predicted defective serine biosynthesis pathway can be rescued with a promiscuous serine hydroxymethyltransferase. Strain CF grew without added thiamine although the thiamine biosynthesis pathway is predicted to be absent; intracellular thiamine diphosphate, the cofactor of carboxylases in central metabolism, was not detected in cell extracts. Thus, strain CF may use amino acids to replenish central metabolites, portending entangled metabolite exchanges in ACT-3. Consistent with annotation, strain CF possesses a functional corrinoid biosynthesis pathway, demonstrated by increasing corrinoid content during growth and guided cobalamin biosynthesis in corrinoid-free medium. Chloroform toxicity to corrinoid-producing methanogens and acetogens may drive the conservation of corrinoid autotrophy in Dehalobacter strains. Heme detection in strain CF cell extracts suggests the ‘archaeal’ heme biosynthesis pathway also functions in anaerobic Firmicutes. This study reinforces the importance of incorporating enzyme promiscuity and cofactor availability in genome-scale functional predictions and identifies essential nutrient interdependencies in anaerobic dechlorinating microbial communities.
Similar content being viewed by others
Introduction
Halogenated organic compounds include a wide array of naturally occurring and man-made chemicals with moderate to high toxicity. Many make excellent hydrophobic solvents. For example, chloroform (CF) and 1,1,1-trichloroethane (1,1,1-TCA) are frequent groundwater contaminants because of their widespread industrial use as solvents and historically poor disposal practices (Furukawa et al., 2005). CF is also one of the most abundant organohalides in the atmosphere (estimated annual global flux is from 0.7 to 4 million t) (Harper, 2000; Cappelletti et al., 2012). In fact, CF in the environment is primarily of natural origin (~90%; McCulloch, 2003), produced by algae in marine systems (Nightingale et al., 1995; Scarratt and Moore, 1999) and by termites in terrestrial systems (Khalil et al., 1990; Laturnus et al., 2002). In addition to neurotoxicity and carcinogenic activity, CF and 1,1,1-TCA are also strong inhibitors of many microbial processes essential for biogeochemical cycling, such as methanogenesis, acetogenesis and reductive dechlorination of chlorinated ethenes (Bagley et al., 2000; Duhamel et al., 2002), and are ozone-depleting substances (Slaper et al., 1996).
Members of the Dehalobacter genus are anaerobes found in sediments of river and marine ecosystems (Zanaroli et al., 2010) as well as groundwater aquifers (Grostern and Edwards, 2006). Dehalobacter are specialized in organohalide respiration (Holliger et al., 1998) or organohalide fermentation (Justicia-Leon et al., 2012; Lee et al., 2012), and are active participants in the global halogen cycle. Based on 16S ribosomal RNA (rRNA) gene sequences (>99% average nucleotide identity), all characterized strains described to date belong to the type species Dehalobacter restrictus and use H2 or formate as an electron donor to reductively dechlorinate a variety of chlorinated and brominated hydrocarbons (Holliger et al., 1998; Sun et al., 2002; Grostern and Edwards, 2009; Yoshida et al., 2009; Grostern et al., 2010; Deshpande et al., 2013; Tang and Edwards, 2013; Nelson et al., 2014; Wang et al., 2014). D. restrictus strains CF and DCA were enriched from a mixed culture referred to as ACT-3 that dechlorinates 1,1,1-TCA via 1,1-dichloroethane (1,1-DCA) to chloroethane (CA) and CF to dichloromethane (DCM). ACT-3 is also used commercially for bioaugmentation (http://sirem-lab.com/products/kb-1). Reductive dehalogenases (RDases) are corrinoid-dependent enzymes that catalyze organohalide dehalogenation (Jugder et al., 2016). In ACT-3, the dechlorination of CF and 1,1,1-TCA is catalyzed by CfrA (CF reductase subunit A) from strain CF; the dechlorination of 1,1-DCA is catalyzed by DcrA from strain DCA. CfrA and DcrA were the only two RDases found expressed of the 19 RDase genes in ACT-3 (Tang and Edwards, 2013).
As organohalide-respiring bacteria like Dehalobacter are key factors in sustaining the global halogen cycle (Krzmarzick et al., 2012) and in bioremediation (Jackson, 2004; Justicia-Leon et al., 2012; Jugder et al., 2016), realizing their metabolic and physiological characteristics is of great importance. This characterization enables more accurate activity predictions, as illustrated by a recent study that found that Dehalococcoides, another genus of organohalide-respiring bacteria, requires a specific form of corrinoid cofactor to degrade the carcinogen vinyl chloride (Yan et al., 2015). The closed genomes of strains CF and DCA were previously assembled from the metagenome of the ACT-3 mixed culture (Tang et al., 2012) and available under NCBI accession numbers CP003869 and CP003870. These two highly similar genomes (>90% whole-genome average nucleotide identity) were then compared with three other available Dehalobacter genomes (strains PER-K23, E1 and UNSWDHB) to categorize common and unique genome-inferred physiological and metabolic traits (Tang et al., 2016). The genomes differed primarily in the complement of RDases and the completeness of the corrinoid biosynthesis pathway. However, all five genomes harbor the nearly identical complement of genes ascribed to central metabolism (the TCA cycle) and amino-acid and cofactor biosynthesis. All strains appear to have the same defects in serine, biotin and thiamine biosynthesis, whereas they all harbor a complete archaeal heme biosynthesis pathway (Tang et al., 2016).
Recent advances in bioinformatics and sequencing technology have generated a large number of genomes and metagenomes from a wide variety of environments that are being mined for functional and ecological traits. However, bioinformatic analyses, while powerful, are often insufficient to reliably predict many metabolic features of an organism because of the considerable number of mis-annotations and hypothetical genes in every genome (Sañudo-Wilhelmy et al., 2014). A relevant example is the recent discovery of the anaerobic biosynthesis genes of 5,6-dimethylbenzimidazole (DMB; Hazra et al., 2015), the lower ligand of the best known corrinoid, cobalamin (vitamin B12). The genes encoding 5-hydroxybenzimidazole synthase (bzaABF) were previously wrongly annotated as thiamine biosynthesis genes because of the homology-based annotation (Hazra et al., 2015).
Here we describe the isolation, growth, morphological characteristics, and electron donor and electron acceptor substrate ranges of strains CF and DCA. Guided by previous genomic and transcriptomic analyses (Rupakula et al., 2013; Tang et al., 2016), we experimentally tested specific genomic predictions, particularly where annotations were uncertain or inconsistent with the observed phenotypes. Tang et al. (2016) classified and curated genome annotations of five Dehalobacter strains including strains CF and DCA. As the orthologous proteins from the five strains are highly similar, typically sharing >98% protein sequence identity, strain CF was chosen as an appropriate model organism for pathways common to all sequenced strains. We explored the predicted gaps in the TCA cycle, potential defects in amino-acid biosynthesis and were able to refine annotations of genes involved in serine biosynthesis. We also evaluated functionality of biosynthetic pathways for biotin, corrinoid, heme and thiamine. These experimental efforts verify genome annotations and bring previous proteomic data of ACT-3 and strain PER-K23 cultures (Rupakula et al., 2013; Tang and Edwards, 2013) into context.
Materials and methods
Isolation and growth conditions
For strain isolation, two highly enriched subcultures, Dehalobacter-1,1,1-TCA/H2 and Dehalobacter-1,1-DCA/H2, were constructed by feeding transfers from the parent mixed culture ACT-3 (Tang and Edwards, 2013) with either 1,1,1-TCA or 1,1-DCA (2 mM nominal) as the electron acceptor in minimal mineral medium (MM medium; Edwards and Grbić-Galić, 1994). H2 was provided as the electron donor (25% in the headspace; 20 mM nominal). Sodium acetate (5 mM) was provided as the carbon source. Specifically, for dilution-to-extinction transfers, inside an anoxic chamber, 7 ml of MM medium, 3 ml of ACT-3 mixed culture supernatant (described in Supplementary Information), sodium acetate (5 mM), 2 μl of neat chlorinated substrate (1,1,1-TCA or 1,1-DCA), and 6.4 ml of H2/CO2 (80% v/v) were added to a 25 ml butyl rubber stopper-sealed and crimped Bellco glass tube. After autoclaving, 10 μl of 1000x filter-sterilized vitamin solution (Supplementary Information) and 20 μl of autoclave-sterilized iron sulfide (FeS) slurry (~8 g l–1; Supplementary Information) were injected into each tube. The enriched subcultures were serially diluted from 10−1 to 10−9. To minimize carryover, inocula were pelleted down to remove supernatant, and were resuspended in and equal volume of sterile MM medium before inoculation. The diluted transfers were incubated at room temperature and dechlorination was monitored by gas chromatography as described in Supplementary Information. Growth conditions for experiments evaluating electron donors, electron acceptors and vitamin requirement were similar to the conditions for these dilution-to-extinction transfers with some modifications (Supplementary Information). The average volumetric dechlorination rates of cultures were determined from the total amount of dechlorinated products generated per day in each bottle divided by the culture volume when >90% of the chlorinated substrates were depleted in the three replicates.
Isolate purity tests
Contamination of the isolate cultures with other microorganisms was investigated by making transfers (10−3 dilution) into MM medium described above but excluding the chlorinated substrates (to detect homoacetogens), or replacing chlorinated electron acceptors with 5 mM NaNO3 (for nitrate reducers), 5 mM NaSO4 (for sulfate reducers), a mixture of methanol, ethanol and lactate (for fermenting microbes), or 0.1 g l–1 yeast extract. These new transfers were incubated at room temperature for 1 month and then examined by restriction fragment length polymorphism (Supplementary Information) and epifluorescence microcopy with 4′,6-diamidino-2-phenylindole dihydrochloride staining (to look for any growth) using standard protocols as described in Supplementary Information. Two pairs of specific primers targeting cfrA and dcrA (Supplementary Information) developed previously (Tang and Edwards, 2013) were used to examine the potential cross-contamination of strains CF and DCA in their isolate cultures.
Enzyme activity assays
Established assays for malate dehydrogenase, succinate dehydrogenase, NADP-dependent malic enzyme (NDME), and RDase activity were conducted using cell extracts prepared from Dehalobacter strain CF and from E scherichia coli strain K-12 (Supplementary Information). Cell extracts from both cultures were adjusted to a final total protein concentration of 50 μg ml–1 in each assay. Serine hydroxymethyltransferase (SHMT) and threonine aldolase activity assays were conducted using purified recombinant SHMT from strain CF (accession number AFV06891). Cloning, expression and purification of strain CF SHMT is described in Supplementary Information. Protein concentrations of cell extracts were determined using the Pierce bicinchoninic acid protein assay kit (Thermo Fisher Scientific, Mississauga, ON, Canada) with a linear range spanning from 20 to 2000 μg ml–1 of protein. Specific activity is reported as μmol of substrate consumed or product generated per min per mg of protein, as well as in ncat mg–1.
Extraction and derivatization of intracellular thiamine species
Thiamine can exist as free and phosphorylated forms within a cell. To measure the concentration of intracellular thiamine species, a protocol involving derivatization into thiochromes was used (Supplementary Information; Leonardi and Roach, 2004).
Corrinoid purification, measurement and structure characterization
Corrinoid content in strain CF culture was quantified using a Lactobacillus growth-based B12 assay following established protocols with some modifications (Yan et al., 2012). For the B12 assay, strain CF cells grown in corrinoid-free medium were transferred twice (10−2 dilution) to exclude any carryover. The third transfer was sampled over time during growth and the samples were assayed using the growth-based Lactobacillus auxotroph assay. In this assay, Lactobacillus growth increased linearly over a corrinoid concentration range from 2 to 20 ng l–1 (Supplementary Figure S1). To determine total corrinoid content in cultures, pH of the samples was adjusted to 5–6 with 3% acetic acid, and the samples were incubated in a boiling water bath for 30 min, followed by centrifugation at 16 000 g for 15 min at 4 °C. The supernatants were diluted 10–50 times and stored at −20 °C until use. The corrinoids produced by strain CF were extracted and derivatized following previous established protocols (Yan et al., 2012), and characterized using ultra performance liquid chromatography-mass spectroscopy (Supplementary Information).
Analytical procedures
Gas chromatography to measure concentrations of chlorinated substrate analyses, high pressure liquid chromatography for measuring thiamine derivatives and acetaldehyde, and ultra performance liquid chromatography-mass spectroscopy for biotin, corrinoid, heme and amino-acid detection are described in the Supplementary Information.
Results and Discussion
Isolation and characterization of Dehalobacter strains
Isolation and verification of purity
Isolation of the strains CF and DCA was achieved via repeated dilution-to-extinction transfers from the original mixed parent culture, ACT-3. The addition of autoclaved sterile mixed culture supernatant prepared from the ACT-3 parent culture was required to sustain the growth of the 10−9 dilution transfers (that is, detection of dechlorination activity) (Supplementary Figure S2). Nineteen successive 10−9 dilution transfers were performed. To replace mixed culture supernatant in the growth medium for the isolate cultures, a chemically defined medium (Supplementary Information) similar to that used for the isolation of D. restrictus strain PER-K23 (Holliger et al., 1998) was tested. Cultures of the two isolates were able to deplete 1 mM chlorinated substrate (1,1,1-TCA or 1,1-DCA) after three successive 10% transfers (v/v) without mixed culture supernatant in MM medium when 0.1 mM of each arginine, histidine and threonine were provided (Figure 1a). These three amino acids may not represent the only viable combination required for growth but other combinations were not explored. The transfer cultures of the two isolate strains stopped dechlorinating when any of the three amino acids was excluded from the medium (data not shown). Based on the highly similar 16S rRNA gene sequences of strains CF and DCA to strain PER-K23 (99.3% average nucleotide identity), and their shared carbon source requirement, strains CF and DCA belong to the type species D. restrictus.
Dechlorination over three serial 10% transfers of strain CF. (a) cultures grown on chemically defined medium amended with the three vitamins (biotin, cobalamin and thiamine see Supplementary Information for exact composition). (b) cultures grown in medium lacking biotin, cobalamin and thiamine. (c) average dechlorination rates for strain CF cultures grown on medium with different vitamin combinations noted below. Symbols: ♦, 1,1,1-TCA; ◊, 1,1-DCA. Data are mean±s.e. of three replicates in each experiment.
Microscopy suggested no growth in chlorinated substrate-free transfer cultures amended with yeast extract, a mixture of methanol, ethanol and sodium lactate, or with electron acceptors nitrate and sulfate. To corroborate the purity of isolates, DNA was extracted from isolate cultures and the 16S rRNA gene was amplified by general bacterial primers 27f and 1541r. Restriction fragment length polymorphism patterns reflected the presence of the only two Dehalobacter 16S rRNA genes in the isolate cultures (Supplementary Figures S3A-C). Strain CF cultures did not dechlorinate 1,1-DCA, and strain DCA cultures did not dechlorinate CF and 1,1,1-TCA. Moreover, cfrA was only amplified from strain CF DNA, and dcrA was only amplified from strain DCA DNA (Supplementary Figure S3D), confirming that the two D. restrictus strains were indeed distinct.
Morphology
Microscopic images of samples of both isolate cultures stained with 4′,6-diamidino-2-phenylindole dihydrochloride revealed a uniform cellular morphology, further supporting the culture purity (Figures 2a and b). Consistent with the three previously described Dehalobacter isolates (strains PER-K23, TCA1 and UNSWDHB; Holliger et al., 1998; Sun et al., 2002; Wong et al., 2016), the cells of strains CF and DCA are either straight or curved rods (Figures 2a and b) approximately 2–6 μm long with a diameter of 0.4–0.6 μm (Figure 2c). Similar to strain PER-K23 (Holliger et al., 1998), strain CF stains Gram-negative (Supplementary Figure S4), whereas it is phylogenetically Gram-positive. This negative staining may result from the presence of a proteinaceous S-layer on the exterior surface of Dehalobacter cells (Figure 2d; Holliger et al., 1998). In some strain CF cells, an electron-dense granule was also observed (Figure 2d).
Microscopic images of Dehalobacter strains. Epifluorescence microscopic images of strains CF (a) and DCA (b) at x1000 magnification. (c) Scanning electron microscopic image and (d) transmission electron microscopic image of the strain CF cell. Scale bars are shown in each panel. Note the long rods in Figure 2b are formed by multiple strain DCA cells lining up end-to-end. CM, the cytoplasmic membrane; G, the electron-dense granule; P, the peptidoglycan layer; S, the proteinous S-layer.
Electron donors and acceptors
According to annotations, Dehalobacter genomes possess various oxidoreductase genes involved in fermentation (Tang et al., 2016), suggesting that they may be able to use electron donors other than H2 or formate. The only genes for electron-accepting reactions in anaerobic respiration identified in the genomes were RDases. A broad range of electron donors and acceptors were tested to evaluate the substrate spectra of the both strains CF and DCA. Of the many electron donors tested, the two strains were only able to oxidize H2 and formate (Supplementary Table S1A). The ability of both strains to use formate as the electron donor is consistent with the expression of Dehalobacter formate dehydrogenase (EC 1.2.1.2) in both 1,1,1-TCA- and 1,1,-DCA-fed ACT-3 mixed cultures (Tang and Edwards, 2013). However, the ability to use formate appeared to be conditional on the addition of ACT-3 mixed culture supernatant, yeast extract or casamino acids to the MM medium. Dechlorination with formate as electron donor was not sustained in medium that was not supplemented (data not shown). This conditional formate utilization may result from the inability to synthesize molybdopterin, the cofactor for formate dehydrogenase. Genome annotation previously revealed that none of the strains possess complete molybdopterin biosynthesis genes (Tang et al., 2016). Therefore, strains CF and DCA may rely on molybdopterin supplementation from other community members in ACT-3 to respire formate. Unfortunately, the absence of commercially available molybdopterin standards has hampered our attempt to study this further.
We determined that strain CF could use 1,1,2-trichloroethane (1,1,2-TCA) as electron acceptor in addition to 1,1,1-TCA and CF. Strain DCA could also use 1,1,2-TCA in addition to 1,1-DCA (Supplementary Table S1B). However, the tolerance of strain CF to aqueous CF concentrations ([CF]aq) is much lower (below 0.42 mM) than to [1,1,1-TCA]aq and [1,1,2-TCA]aq (above 1 mM). The mixed culture was much less sensitive to CF, tolerating above 1 mM. No other chlorinated ethene or ethane tested was dechlorinated by the isolates (Supplementary Table S1B). As we could amend with higher concentrations, 1,1,1-TCA was used as the electron acceptor for strain CF in experiments to verify genome annotations. The genomes of strains CF, DCA and UNSWDHB contain a complete operon (pceABCT) in their genomes encoding an ortholog of the characterized tetrachloroethene (PCE) RDase PceA from strain PER-K23 (Tang et al., 2016; Wong et al., 2016) (>89% amino-acid identity). However, none of the strains was found to dechlorinate PCE. This lack of activity may result from the loss of a functional transcription factor to induce PceA expression or from a mutation at key catalytic residues.
To determine if CfrA can also dechlorinate 1,1,2-TCA, we conducted RDase enzyme assays using cell extracts of pure strain CF culture grown on 1,1,1-TCA that is known to express only CfrA. These cell extracts not only dechlorinated 1,1,1-TCA, but also CF and 1,1,2-TCA, confirming substrate promiscuity of CfrA (Table 1). Heat-treated cell extracts did not catalyze 1,1,2-TCA dechlorination. Moreover, the fact that strain DCA is able to respire 1,1,2-TCA, and that DcrA is the only RDase detected in 1,1-DCA grown cells (Tang and Edwards, 2013) suggest DcrA also dechlorinates 1,1,2-TCA. 1,1,2-TCA is therefore a common substrate for four similar RDases, including CfrA and DcrA described above, TmrA from strain UNSWDHB (Wong et al., 2016), and CtrA from Desulfitobacterium sp. strain PR (Zhao et al., 2015). These chlorinated alkane-dechlorinating RDases define a protein cluster (ortholog group) based on >90% amino-acid identity (Hug and Edwards, 2013; Tang et al., 2016). Interestingly, each strain and corresponding active RDase have slightly different substrate preferences supporting a previous contention that differentiation of organohalide-respiring strains is driven by chlorinated substrates (Islam et al., 2010; Tang et al., 2016).
Experimental verification of genome annotations of strain CF
Missing TCA cycle genes
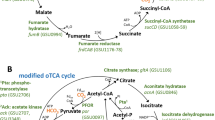
The TCA cycle in all five sequenced Dehalobacter genomes is predicted to be incomplete because the genes encoding malate dehydrogenase and succinate dehydrogenase were not found (Figure 3, pathways marked with red x). If these genes are truly absent, neither fumarate nor malate could be synthesized. Dehalobacter strains need the TCA cycle to synthesize oxaloacetate and 2-oxoglutarate for amino-acid biosynthesis (Miflin and Lea, 1977). The absence of the two enzymes may be associated with the amino-acid requirement of Dehalobacter. Alternatively, a gene annotated as NDME (EC 1.1.1.40) is present in all Dehalobacter genomes and may have a role in compensating for missing TCA cycle genes. If active, this predicted NDME gene product could catalyze pyruvate–malate conversion (Figure 3). Moreover, the annotated malic enzyme is also expressed in the ACT-3 mixed culture and proteome of strain PER-K23 (Rupakula et al., 2013, Tang and Edwards, 2013). We assayed cell extracts of strain CF and E. coli (positive control) for malate and succinate dehydrogenase as well as NDME activities (Table 2). The latter activity was detected in both strain CF (0.06 μmol min–1mg–1 or 1 nkat mg–1) and E. coli (0.01 μmol min–1mg–1 or 0.17 nkat mg–1), whereas malate dehydrogenase and succinate dehydrogenase activities were only detected in E. coli cell extracts. Consistent with annotation, enzyme assays revealed that the activities of malate and succinate dehydrogenase are absent in Dehalobacter genomes. However, the NDME can salvage the synthesis of malate and fumarate from pyruvate (Figure 3).
Schematic of the proposed metabolic map of strain CF. The red crosses (X) represent missing genes in the strain CF genome, whereas a red curved dashed line beside a reaction represents the absence of the essential cofactor. The black arrows represent nutrients imported from medium. Note that the threonine–glycine conversion (highlighted in blue) is catalyzed by promiscuous SHMT. Abbreviations for cofactors and enzymes involved in the reactions are listed below with full name: CfrA, chloroform reductive dehalogenase; PLP, pyridoxal phosphate; TDP, thiamine diphosphate; PK, pyruvate kinase; PPDK, pyruvate phosphate dikinase; FDH, formate dehydrogenase (molybdopterin); FHL, formate hydrogen lyase complex (molybdopterin); ACS, acetate:CoA ligase (AMP-forming); PFL, pyruvate formate-lyase; PFO, pyruvate-flavodoxin oxidoreductase (TDP); PCT, pyruvate carboxyl transferase (biotin); NDME, NADP-dependent malic enzyme (NADP); CS, citrate synthase; AH, aconitate hydratase; ICDH, isocitrate dehydrogenase (NADP); 2-OGs, 2-oxoglutarate:ferredoxin oxidoreductase (TDP); SCL, succinyl-CoA ligase; FH, fumarate hydratase; GK, glucokinase; PSP, phosphoserine phosphatase; OAD, oxaloacetate decarboxylase (biotin); SDH, succinate dehydrogenase(heme); SDHT, serine dehydratase; MDH, malate dehydrogenase; ICL, isocitrate lyase; MS, malate synthase; TAL, threonine aldolase (PLP); SHMT, serine hydroxymethyltransferase (PLP). Other abbreviations: CM, cytoplasmic membrane; CW, cell wall; Cyt b, cytochrome b; WL pathway, Wood–Ljungdahl pathway.
Serine biosynthesis
Strains CF, DCA and PER-K23 were found to require arginine, histidine and threonine for growth, yet the biosynthesis pathways for these three amino acids appear complete in all the five Dehalobacter genomes. Rather, their genomes are missing the gene encoding phosphoserine phosphatase (PSP; EC 3.1.3.3), the enzymes that catalyzes the last step of the conventional serine biosynthesis pathway (Greenberg and Ichihara, 1957), whereas other serine salvage pathways are not present (Supplementary Figure S5A). A defect in serine biosynthesis should disable biosynthesis of glycine and cysteine (Supplementary Figure S5). However, strains CF and DCA are able to grow in a defined medium (supplemented with arginine, histidine and threonine; Figure 3) without cysteine, glycine and serine, suggesting the presence of an alternative mechanism to produce serine, most likely from the three amino acids provided, as explained below.
Threonine can be converted to serine via glycine (Figure 3). However, threonine aldolase (EC 4.1.2.5), the enzyme catalyzing the threonine–glycine conversion, is absent in the five Dehalobacter genomes. Interestingly, SHMT (EC 2.1.2.1), the enzyme catalyzing glycine–serine conversion, has been reported to possess promiscuous threonine aldolase activity in both E. coli and Methanosarcina barkeri strain Fusaro (Ogawa et al., 2000; Buchenau and Thauer, 2004). SHMT is also expressed in the proteome of strain PER-K23 (Rupakula et al., 2013). Furthermore, the SHMT of M. barkeri strain Fusaro shares >59% protein sequence identity to the sequences of the five Dehalobacter SHMTs, indicating Dehalobacter may utilize SHMT to synthesize serine from threonine. The lack of a PSP-coding gene can also be rescued by other promiscuous phosphatases in the haloacid dehalogenase (HAD) family. For example, it has been reported that histidinol phosphatase (HisB; EC 3.1.3.15) in histidine biosynthesis can rescue missing PSP in E. coli strain K-12 and Thermococcus onnurineus strain NA1 (Patrick et al., 2007; Lee et al., 2008; Yip and Matsumura, 2013). However, histidinol phosphatases (HisJ) in Dehalobacter genomes does not belong to the HAD family (Tang et al., 2016).
To test if Dehalobacter can synthesize serine, the gene for SHMT (accession number AFV06891) from strain CF was cloned, heterologously expressed and purified (Supplementary Figure S5B). Strain CF SHMT revealed both threonine aldolase activity (0.07 μmol min–1mg–1 or 1.2 nkat mg–1) and hydroxyl methyltransferase activity (3.4 μmol min–1mg–1 or 57 nkat mg–1; Table 2), suggesting that strain CF can synthesize serine from threonine via glycine. None of the enzyme-free or substrate-free controls showed any activity. Thus, the presence of a serine salvage mechanism enables serine production from threonine (also from glycine or cysteine) in strain CF. However, this does not explain why strain CF requires threonine as the biosynthesis pathway for threonine is complete. This discrepancy is further discussed in the section on thiamine section below.
In a recent comparative genomic analysis of >6000 sequenced bacteria from diverse environments, most Firmicutes were found to be serine auxotrophs based on annotated defects in the serine biosynthesis pathway (Mee et al., 2014), consistent with previous annotation of Dehalobacter genomes (Tang et al., 2016). However, we have now shown that these ostensible serine auxotrophs can actually synthesize serine using promiscuous enzymes, illustrating the shortcomings of typical bioinformatic analyses without experimental verification. Models constructed based on this comparative genomic analysis may assume that Firmicutes require serine from other community members when in fact they do not. For example, Desulfitobacterium hafniense strain Y51, another organohalide-respiring model organism and a member of Firmicutes, may also synthesize serine using promiscuous SHMT because (i) the organism lacks the PSP-coding gene in its genome (Nonaka et al., 2006, ii) its SHMT (accession number BAE86716) shares 75% protein sequence identity with the homolog in strain CF and (iii) our data revealed that D. hafniense strain Y51 can grow in defined medium without amino-acid supplementation (Bi, 2015).
Biosynthesis of organic cofactors
The growth reliance of Dehalobacter on the three amino acids can also result from defects in cofactor biosynthesis. It is possible that (i) Dehalobacter cannot synthesize specific cofactors or (ii) Dehalobacter is not able to transform certain cofactor pre-cursors into functional forms. For example, thiamine is converted to thiamine monophosphate or thiamine diphosphate (TDP) intracellularly to be functional (Leonardi and Roach, 2004). In genomes of strains CF and DCA, the biosynthetic pathway for biotin appears to be missing 4/5 genes and for thiamine the pathway is missing 5/9 genes (Tang et al., 2016). To determine if these pathways are truly incomplete, growth of strain CF was tested in the absence of these cofactors. We also tested the growth of strain CF in the presence and absence of cobalamin as genome annotation predicted a complete corrinoid biosynthesis pathway. Growth of strain CF was sustained in the absence of these three cofactors (biotin, cobalamin and thiamine) in cultures that were spun down and transferred three successive times (10% v/v inoculum; Figure 1b). Removal of cobalamin decreased the dechlorination rate by about a factor of 3 (from 30 to 10 μM per day nominal), whereas exclusion of biotin and thiamine from the medium did not affect the growth rate (11 μM per day nominal; Figure 1c). When grown with the three vitamins, the cell yield for strain CF determined from microscopic cell counts, which is 5.73 (±1.79) × 1012 (n=9) cells/mol Cl− released, is comparable to the qPCR-determined cell yield of strain CF in CF-fed mixed culture ACT-3, which is 6 (±0.03) × 1012/mol Cl− released (Grostern et al., 2010).
Biotin biosynthesis and metabolism
Biotin is considered vital for organisms in all domains of life because of its essential role in fatty acid biosynthesis (cofactor for acetyl-CoA carboxylase; Lin and Cronan, 2011), yet no known biotin biosynthesis pathway was identified in the five Dehalobacter genomes even though both strain PER-K23 (Holliger et al., 1998) and strain CF (Figure 1c) can grow without added biotin. It was reported that biotin-independent malonate decarboxylases (Hoenke et al., 1997) can circumvent the need for biotin in fatty acid biosynthesis; however, a homolog of this gene was not found in the five Dehalobacter genomes. Biotin is also an essential cofactor for pyruvate carboxyl transferase (Figure 3) that can convert pyruvate to oxaloacetate for amino-acid biosynthesis, and the pyruvate carboxyl transferase coding gene is annotated in all the five Dehalobacter genomes (Tang et al., 2016). Moreover, traces of biotin may be sufficient to support growth. It was reported that as few as 1000 biotin carboxyl carrier proteins are present in each E. coli cell (Li and Cronan, 1993). Accordingly, medium with a biotin concentration of 0.4 μg l–1 should be enough to support the growth of an E. coli culture to a cell density of 109 cells ml–1.
To determine if biotin is present at sufficient quantities in our medium as a contaminant, we analyzed samples of the concentrated stock solutions used to make the MM medium by liquid chromatography-mass spectroscopy. We detected biotin in the 1000x vitamin stock solution (excluding biotin; referred to as biotin−) at a concentration of 15 μg l–1 (Supplementary Figure S6), indicating that (after 1000x dilution of the stock) the medium may contain ~15 ng l–1 biotin. This amount is low but may be sufficient to support the growth of strain CF cultures because they only reach a cell density ~108 cells ml–1 and require biotin for anabolism but not for respiration. Nevertheless, we cannot exclude the possibility that an uncharacterized biotin biosynthesis pathway is functioning in strain CF.
Thiamine biosynthesis and metabolism
The absence of a thiamine biosynthesis pathway in Dehalobacter genomes is consistent with the observation that the growth of strain PER-K23 required thiamine supplementation (Holliger et al., 1998). In contrast, the growth of strain CF seems not to require thiamine addition (Figure 1c). TDP is the cofactor of pyruvate-flavodoxin oxidoreductase and 2-oxoglutarate decarboxylase, two important carboxylases in central metabolism (Figure 3; Pohl et al., 2002). To test whether strain CF can synthesize TDP from thiamine, cultures of strain CF and E. coli grown on thiamine-containing medium (25 μg l–1) were harvested for intracellular thiamine derivatives analysis. TDP was only detected in E. coli cell extracts, whereas thiamine was detected in both strain CF and E. coli cell extracts (Figure 4). Interestingly, although a gene annotated as thiamine pyrophosphokinase (EC 2.7.6.2) is present in the strain PER-K23 genome (Tang et al., 2016), the enzyme was not found to be expressed in the strain PER-K23 proteomes (Rupakula et al., 2013), consistent with the lack of TDP detection.
Determination of intracellular thiamine derivatives in strain CF cells. Chromatograms from high pressure liquid chromatography (HPLC) analysis with fluorescence detection (excitation at 365 nm and emission at 435 nm) of: (a) 1 μM thiamine standard solution; (b) 1 μM thiamine monophosphate; (c) 1 μM thiamine diphosphate; (d) cell extract of E. coli; and (e) cell extract of strain CF.
The lack of TDP indicates that strain CF only uses pyruvate formate-lyase for acetyl-CoA–pyruvate conversion, and not the TDP-dependent pyruvate-flavodoxin oxidoreductase (Figure 3). However, it is unclear if an organism could use only pyruvate formate-lyase to assimilate acetate and formate into pyruvate in defined medium. The only relevant study we found in literature did not test the effect of deleting the pyruvate-flavodoxin oxidoreductase-like ydbK (Nakayama et al., 2013) in their E. coli strain to exclude its contribution to pyruvate production (Zelcbuch et al., 2016). Moreover, threonine can be converted into pyruvate via serine (Figure 3). Furthermore, strain CF cultures grown on pyruvate as the main carbon source dechlorinated at twice the rate (53 μM per day nominal) of cultures grown on acetate (29 μM per day nominal; amino acids supplemented in both conditions; Supplementary Figure S7). Therefore, strain CF likely requires amino-acid supplementation for growth partially because of difficulties in acetyl-CoA–pyruvate conversion, which in turn block synthesis of oxaloacetate and thus 2-oxo-glutarate (pre-cursors for amino acids; Figure 3; Miflin and Lea, 1977). Such nutrient requirements for strain CF portend a tightly entangled syntrophy in the ACT-3 microbial community.
Corrinoid biosynthesis
Corrinoids, such as cobalamin, have a crucial role in methionine and ribonucleotide biosynthesis, as well as energy metabolism in organohalide-respiring bacteria (Miller et al., 1998; Maillard et al., 2003; Sañudo-Wilhelmy et al., 2014). Reductive dechlorination and growth of organohalide-respiring bacteria are usually enhanced with corrinoid supplementation (Yan et al., 2013). It is reported that strains TCA1 (Sun et al., 2002) and UNSWDHB (Wong et al., 2016) grow without corrinoid supplementation. Strain CF could also grow without corrinoid supplementation (Figure 1b). Consistent with these observations, genomes of strains CF, DCA (Tang et al., 2016) and UNSWDHB (Deshpande et al., 2013) all possess a complete cobinamide biosynthesis pathway, whereas strains PER-K23 (Rupakula et al., 2013) and E1 (Maphosa et al., 2012) do not. However, to date, no direct evidence for de novo corrinoid synthesis by Dehalobacter exists. Thus, the corrinoid concentration in strain CF cultures grown in corrinoid-free medium was quantified using a microbial B12 assay (Yan et al., 2013). The significant increase of corrinoid concentration along with dechlorination confirms the ability of strain CF to synthesize corrinoid de novo (Figures 5a and b). We attempted to characterize the type of corrinoid using UPLC-ESI-MS, however, none of the known cobamides with a benzimidazolyl or phenyl type lower ligand was detected in the cell extract of strain CF culture grown on corrinoid-free medium based on m/z values and retention times of the known cobamide standards (Supplementary Table S2). Nevertheless, cobalamin was identified in cell extracts of strain CF culture when 10 μM of the lower ligand compound DMB was supplemented to the corrinoid-free medium (Figure 5c). The guided cobalamin biosynthesis confirmed that strain CF possesses a functional cobinamide biosynthesis pathway to assemble cobalamin at the presence of DMB (Tang et al., 2016). No cobinamide or cobalamin was identified in the commercial DMB and medium used for growth (Supplementary Figure S8). Thus, strain CF produces a cobamide that is yet to be identified, likely a purine species as the lower ligand as reported for strains of Clostridium (adenine) and Desulfovibrio species (guanine and hypoxanthine) (Hoffmann et al., 2000; Guindon and Gascuel, 2003).
Corrinoid production and guided cobalamin synthesis by strain CF. (a) Dechlorination profiles (production of 1,1-DCA from 1,1,1-TCA) and (b) corresponding corrinoid production in cultures grown under different conditions as shown. ●, autoclaved negative control (NC); ■, 1,1,1-TCA-free control (TCA−); ∆, cobalamin- and DMB-free cultures (Cbl−); ▼, corrinoid-free and DMB-containing cultures (DMB+); ○, cobalamin-containing positive control (Cbl+). (c) UPLC-ESI-MS analysis of guided cobalamin synthesis in strain CF culture with DMB supplementation. The chromatograms on top panel shown are extracted ion chromatograms (m/z=1355.57±0.013) and the chemicals are ionized in positive mode. Bottom panel shows mass spectrum. CN-Cbl STD, cyanocobalamin standard. Data are mean±s.e. of three replicates in each experiment.
None of the bza genes involved in anaerobic DMB biosynthesis (Hazra et al., 2015) was identified in the five Dehalobacter genomes (Tang et al., 2016), consistent with the observation that no benzimidazole-type cobamide was identified from the strain CF culture. However, when DMB was provided, strain CF preferentially synthesized cobalamin rather than its native cobamide. This is in agreement with the discovery that DMB is a universally preferred substrate for the lower ligand-activating enzyme (CobT) in cobamide biosynthesis (Hazra et al., 2013). Dechlorination rates in DMB-supplemented cultures were comparable to rates in cobalamin-supplemented cultures (Figure 5a), indicating that corrinoid synthesis in strain CF is limited by lower ligand availability but not by the availability of the porphyrin backbone. Furthermore, total corrinoid concentrations are higher in DMB-supplemented cultures than those without DMB (Figure 5b).
The Dehalobacter strains enriched from CF- and 1,1,1-TCA-dechlorinating consortia have retained their ability to synthesize corrinoids, whereas strains PER-K23 and E1 enriched with other halogenated substrates (PCE and β-hexachlorocyclohexane) do not. Strains PER-K23 and E1 do have partial pathways, but specific functions or sections have been lost, perhaps during enrichment. CF and 1,1,1-TCA, or methylchloroform, are potent inhibitors of homoacetogens and methanogens, well-known corrinoid producers in anaerobic environments (Whitman and Wolfe, 1984; Stupperich et al., 1988). In addition, most corrinoid biosynthesis genes are missing from the dominant fermenting microbe, a Bacteroides sp., in ACT-3 (Tang et al., 2012). Strain CF may have conserved its corrinoid-synthesizing ability because of environmental corrinoid shortage, whereas other corrinoid-auxotrophic strains lost the ability as a result of an abundant supply from other community members or enrichment in laboratory medium containing B12. For example, the β-hexachlorocyclohexane-dechlorinating strain E1 has only a partial cobinamide pathway and depends on the syntrophic relationship with a Sedimentibacter sp., which contains a complete cobinamide biosynthesis pathway in its genome (Maphosa et al., 2012). Moreover, PCE-dechlorinating strain PER-K23 possesses complete and functional cobinamide biosynthesis genes, except for a truncated CbiH gene, resulting in its corrinoid auxotrophy (Rupakula et al., 2015). Thus 1,1,1-TCA and CF may indirectly contribute to the conservation of the corrinoid biosynthesis genes among D. restrictus genomes.
Heme biosynthesis
Heme (heme b) is a cofactor for cytochrome b, a subunit of hup-type Ni,Fe-hydrogenase complex found in all five Dehalobacter genomes (Tang et al., 2016). The hup-type Ni,Fe-hydrogenase is expressed during growth of strain PER-K23 (Rupakula et al., 2013) and in all ACT-3 subcultures (Tang and Edwards, 2013), although the membrane-bound cytochrome b subunit was not detected, perhaps because of the limit of tryptic digestion-based proteomics. All five Dehalobacter genomes were found to possess a complete archaeal heme biosynthesis pathway first characterized in the methanogenic archaeon Methanosarcina barkeri (Kuhner et al., 2014; Tang et al., 2016). Recently, another heme biosynthesis pathway in aerobic Firmicutes was characterized (Dailey et al., 2015). However, one of the biosynthesis enzymes, HemY, which converts coproporphyrinogen III to coproporphyrin III is an oxygenase and thus cannot function in strict anaerobic Firmicutes including Dehalobacter. Accordingly, the so-called ‘archaeal pathway’ is still the only characterized pathway for anaerobic heme biosynthesis.
When heme (hematin form; 25 μg l–1) was added to the medium, Dehalobacter strain CF dechlorinated faster than without heme addition (Figure 6a). To confirm that the heme biosynthesis pathway is functional in strain CF, cell extracts of culture grown in heme-free medium were analyzed by UPLC-ESI-MS. A peak with a retention time of 1.5 min and an m/z value of 633.0404 comparable to that of an authentic heme standard (m/z=633.0380) was identified (Figures 6b and c), confirming that strain CF is able to synthesize heme, most likely via the identified archaeal pathway. Before this experiment, heme has never been included in the components used to make growth medium for Dehalobacter. Our experiment indicates that this ‘archaeal pathway’ is not only functioning in methanogenic archaea but also in some anaerobic Firmicutes.
Heme production by strain CF and its influence on dechlorination. (a) Average dechlorination rates of strain CF cultures grown with/out heme supplementation. (b) Ultra performance liquid chromatography-mass spectroscopy (UPLC-MS) analysis of heme b standard (hematin form) and (c) heme in cell extract of strain CF. Chromatograms shown are extracted ion chromatograms (m/z=633.038±0.0063) and the chemicals are ionized in negative mode. The * symbol in Figure 6c is an unknown organic present in strain CF cell extract. Data are mean±s.e. of three replicates in each experiment.
Implications for microbial ecology
In this study, experiments were conducted to refine or confirm predicted annotations in a strict anaerobe. These improved annotations enhance the accuracy of protein annotations, not only for Dehalobacter, but also for many other microbes, especially anaerobes, with similar pathways and genes. The study also explained several metabolic interdependencies in Dehalobacter-containing mixed microbial communities. Consistent with curated genome annotations, CF-respiring D. restrictus strains are capable of de novo synthesis of the corrinoid cofactor of RDases, which is in contrast to other D. restrictus strains where this ability has been lost. This study revealed that all sequenced Dehalobacter to date are capable of synthesizing heme (cofactor of Ni,Fe-hydrogenase) for organohalide respiration. 1,1,1-TCA and CF, as inhibitors of many fermentative or acetogenic corrinoid producers, may indirectly contribute to the conservation of corrinoid biosynthesis genes in Dehalobacter genomes. Experimental verification of genome annotation is a laborious effort, particularly for fastidious microorganisms, yet critical to resolving inconsistencies, discovering new functions, and enabling genome-scale metabolic modeling in microbial communities. This study illustrates two aspects of the interpretation of genome annotation that can improve the correspondence between bioinformatic predictions and the reality: (i) cofactor availability for corresponding metabolic reactions and (ii) the potential for enzyme promiscuity to rescue apparently missing pathways. These findings reinforce the importance of incorporating cofactor availability for corresponding enzymes in genome-wide function prediction as metabolic reactions require that both functional cofactors and corresponding enzymes be present. Moreover, biochemical characterization of poorly studied alternative metabolic pathways is urgently required to improve accuracy of genome annotations. Finally, the example of serine salvage by promiscuous SHMT in strain CF suggests experimental verification is crucial to verify interpretations drawn from bioinformatic analyses.
References
Bagley DM, Lalonde M, Kaseros V, Stasiuk KE, Sleep BE . (2000). Acclimation of anaerobic systems to biodegrade tetrachloroethene in the presence of carbon tetrachloride and chloroform. Water Res 34: 171–178.
Bi M . (2015) Identification of purinyl-cobamide as a novel corrinoid cofactor of tetrachloroethene reductive dehalogenases in Desulfitobacterium spp. Master of Science thesis, University of Tennessee: Knoxville, TN, USA.
Buchenau B, Thauer RK . (2004). Tetrahydrofolate-specific enzymes in Methanosarcina barkeri and growth dependence of this methanogenic archaeon on folic acid or p-aminobenzoic acid. Arch microbiol 182: 313–325.
Cappelletti M, Frascari D, Zannoni D, Fedi S . (2012). Microbial degradation of chloroform. Appl Microbiol Biotech 96: 1395–1409.
Dailey HA, Gerdes S, Dailey TA, Burch JS, Phillips JD . (2015). Noncanonical coproporphyrin-dependent bacterial heme biosynthesis pathway that does not use protoporphyrin. Proc Natl Acad Sci 112: 2210–2215.
Deshpande NP, Wong YK, Manefield M, Wilkins MR, Lee M . (2013). Genome sequence of Dehalobacter UNSWDHB, a chloroform-dechlorinating bacterium. Genome Announc 1: e00720–00713.
Duhamel M, Wehr SD, Yu L, Rizvi H, Seepersad D, Dworatzek S et al. (2002). Comparison of anaerobic dechlorinating enrichment cultures maintained on tetrachloroethene, trichloroethene, cis-dichloroethene and vinyl chloride. Water Res 36: 4193–4202.
Edwards EA, Grbić-Galić D . (1994). Anaerobic degradation of toluene and o-xylene by a methanogenic consortium. Appl Environ Microbiol 60: 313–322.
Furukawa K, Suyama A, Tsuboi Y, Futagami T, Goto M . (2005). Biochemical and molecular characterization of a tetrachloroethene dechlorinating Desulfitobacterium sp. strain Y51: a review. J Ind Microbiol Biotechnol 32: 534–541.
Greenberg DM, Ichihara A . (1957). Further studies on the pathway of serine formation from carbohydrate. J Biol Chem 224: 331–340.
Grostern A, Edwards EA . (2006). A 1,1,1-trichloroethane-degrading anaerobic mixed microbial culture enhances biotransformation of mixtures of chlorinated ethenes and ethanes. Appl Environ Microbiol 72: 7849–7856.
Grostern A, Edwards EA . (2009). Characterization of a Dehalobacter coculture that dechlorinates 1,2-dichloroethane to ethene and identification of the putative reductive dehalogenase gene. Appl Environ Microbiol 75: 2684–2693.
Grostern A, Duhamel M, Dworatzek S, Edwards EA . (2010). Chloroform respiration to dichloromethane by a Dehalobacter population. Environ Microbiol 12: 1053–1060.
Guindon S, Gascuel O . (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704.
Harper DB . (2000). The global chloromethane cycle: biosynthesis, biodegradation and metabolic role. Nat Prod Rep 17: 337–348.
Hazra AB, Tran JL, Crofts TS, Taga ME . (2013). Analysis of substrate specificity in CobT homologs reveals widespread preference for DMB, the lower axial ligand of vitamin B12. Chem Biol 20: 1275–1285.
Hazra AB, Han AW, Mehta AP, Mok KC, Osadchiy V, Begley TP et al. (2015). Anaerobic biosynthesis of the lower ligand of vitamin B12. Proc Natl Acad Sci 112: 10792–10797.
Hoenke S, Schmid M, Dimroth P . (1997). Sequence of a gene cluster from Klebsiella pneumoniae encoding malonate decarboxylase and expression of the enzyme in Escherichia coli. Eur J Biochem/FEBS 246: 530–538.
Hoffmann B, Oberhuber M, Stupperich E, Bothe H, Buckel W, Konrat R et al. (2000). Native corrinoids from Clostridium cochlearium are adeninylcobamides: spectroscopic analysis and identification of pseudovitamin B12 and factor A. J Bacteriol 182: 4773–4782.
Holliger C, Hahn D, Harmsen H, Ludwig W, Schumacher W, Tindall B et al. (1998). Dehalobacter restrictus gen. nov. and sp. nov., a strictly anaerobic bacterium that reductively dechlorinates tetra- and trichloroethene in an anaerobic respiration. Arch Microbiol 169: 313–321.
Hug LA, Edwards EA . (2013). Diversity of reductive dehalogenase genes from environmental samples and enrichment cultures identified with degenerate primer PCR screens. Front Microbiol 4: 341.
Islam MA, Edwards EA, Mahadevan R . (2010). Characterizing the metabolism of Dehalococcoides with a constraint-based model. PLoS Comput Biol 6: e1000887.
Jackson RE . (2004). Recognizing emerging environmental problems: the case of chlorinated solvents in groundwater. Technol Cult 45: 55–79.
Jugder B-E, Ertan H, Bohl S, Lee M, Marquis CP, Manefield M . (2016). Organohalide respiring bacteria and reductive dehalogenases: key tools in organohalide bioremediation. Front Microbiol 7: 249.
Justicia-Leon SD, Ritalahti KM, Mack EE, Löffler FE . (2012). Dichloromethane fermentation by a Dehalobacter sp. in an enrichment culture derived from pristine river sediment. Appl Environ Microbiol 78: 1288–1291.
Justicia-Leon SD, Higgins S, Mack EE, Griffiths DR, Tang S, Edwards EA et al. (2014). Bioaugmentation with distinct Dehalobacter strains achieves chloroform detoxification in microcosms. Environ Sci Tech 48: 1851–1858.
Khalil M, Rasmussen R, French J, Holt J . (1990). The influence of termites on atmospheric trace gases: CH4, CO2, CHCl3, N2O, CO, H2, and light hydrocarbons. J Geophys Res Atmos 95: 3619–3634.
Krzmarzick MJ, Crary BB, Harding JJ, Oyerinde OO, Leri AC, Myneni SC et al. (2012). Natural niche for organohalide-respiring Chloroflexi. Appl Environ Microbiol 78: 393–401.
Kuhner M, Haufschildt K, Neumann A, Storbeck S, Streif J, Layer G . (2014). The alternative route to heme in the methanogenic archaeon Methanosarcina barkeri. Archaea 2014: 327637.
Laturnus F, Haselmann KF, Borch T, Grøn C . (2002). Terrestrial natural sources of trichloromethane (chloroform, CHCl3—an overview. Biogeochemistry 60: 121–139.
Lee HS, Cho Y, Lee J-H, Kang SG . (2008). Novel monofunctional histidinol-phosphate phosphatase of the DDDD superfamily of phosphohydrolases. J Bacteriol 190: 2629–2632.
Lee M, Low A, Zemb O, Koenig J, Michaelsen A, Manefield M . (2012). Complete chloroform dechlorination by organochlorine respiration and fermentation. Environ Microbiol 14: 883–894.
Leonardi R, Roach PL . (2004). Thiamine biosynthesis in Escherichia coli in vitro reconstitution of the thiazole synthase activity. J Biol Chem 279: 17054–17062.
Li SJ, Cronan JE Jr . (1993). Growth rate regulation of Escherichia coli acetyl coenzyme A carboxylase, which catalyzes the first committed step of lipid biosynthesis. J Bacteriol 175: 332–340.
Lin S, Cronan JE Jr . (2011). Closing in on complete pathways of biotin biosynthesis. Mol bioSyst 7: 1811–1821.
Maillard J, Schumacher W, Vazquez F, Regeard C, Hagen WR, Holliger C . (2003). Characterization of the corrinoid iron-sulfur protein tetrachloroethene reductive dehalogenase of Dehalobacter restrictus. Appl Environ Microbiol 69: 4628–4638.
Maphosa F, van Passel MW, de Vos WM, Smidt H . (2012). Metagenome analysis reveals yet unexplored reductive dechlorinating potential of Dehalobacter sp. E1 growing in co-culture with Sedimentibacter sp. Environ Microbiol Rep 4: 604–616.
McCulloch A . (2003). Chloroform in the environment: occurrence, sources, sinks and effects. Chemosphere 50: 1291–1308.
Mee MT, Collins JJ, Church GM, Wang HH . (2014). Syntrophic exchange in synthetic microbial communities. Proc Natl Acad Sci 111: E2149–E2156.
Miflin B, Lea P . (1977). Amino acid metabolism. Annu Rev Plant Biol 28: 299–329.
Miller E, Wohlfarth G, Diekert G . (1998). Purification and characterization of the tetrachloroethene reductive dehalogenase of strain PCE-S. Arch Microbiol 169: 497–502.
Nakayama T, Yonekura S, Yonei S, Zhang-Akiyama QM . (2013). Escherichia coli pyruvate:flavodoxin oxidoreductase, YdbK—regulation of expression and biological roles in protection against oxidative stress. Genes Genetic Syst 88: 175–188.
Nelson JL, Jiang J, Zinder SH . (2014). Dehalogenation of chlorobenzenes, dichlorotoluenes, and tetrachloroethene by three Dehalobacter spp. Environ Sci Technol 48: 3776–3782.
Nightingale PD, Malin G, Liss P . (1995). Production of chloroform and other low molecular-weight halocarbons by some species of macroalgae. Limnol Oceanogr 40: 680–689.
Nonaka H, Keresztes G, Shinoda Y, Ikenaga Y, Abe M, Naito K et al. (2006). Complete genome sequence of the dehalorespiring bacterium Desulfitobacterium hafniense Y51 and comparison with Dehalococcoides ethenogenes 195. J Bacteriol 188: 2262–2274.
Ogawa H, Gomi T, Fujioka M . (2000). Serine hydroxymethyltransferase and threonine aldolase: are they identical? Int J Biochem Cell Biol 32: 289–301.
Patrick WM, Quandt EM, Swartzlander DB, Matsumura I . (2007). Multicopy suppression underpins metabolic evolvability. Mol Biol Evol 24: 2716–2722.
Pohl M, Lingen B, Müller M . (2002). Thiamin-diphosphate-dependent enzymes: New aspects of asymmetric C-C bond formation. Chem A Eur J 8: 5288–5295.
Rupakula A, Kruse T, Boeren S, Holliger C, Smidt H, Maillard J . (2013). The restricted metabolism of the obligate organohalide respiring bacterium Dehalobacter restrictus: lessons from tiered functional genomics. Philos Trans R Soc Lond B Biol Sci 368: 20120325.
Rupakula A, Lu Y, Kruse T, Boeren S, Holliger C, Smidt H et al. (2015). Functional genomics of corrinoid starvation in the organohalide-respiring bacterium Dehalobacter restrictus strain PER-K23. Front Microbiol 5: 751.
Sañudo-Wilhelmy SA, Gómez-Consarnau L, Suffridge C, Webb EA . (2014). The role of B vitamins in marine biogeochemistry. Ann Rev Mar Sci 6: 339–367.
Scarratt M, Moore R . (1999). Production of chlorinated hydrocarbons and methyl iodide by the red microalga Porphyridium purpureum. Limnol Oceanogr 44: 703–707.
Slaper H, Velders GJ, Daniel JS, de Gruijl FR, van der Leun JC . (1996). Estimates of ozone depletion and skin cancer incidence to examine the Vienna Convention achievements. Nature 384: 256–258.
Stupperich E, Eisinger HJ, Krautler B . (1988). Diversity of corrinoids in acetogenic bacteria. P-cresolylcobamide from Sporomusa ovata, 5-methoxy-6-methylbenzimidazolylcobamide from Clostridium formicoaceticum and vitamin B12 from Acetobacterium woodii. Eur J Biochem/FEBS 172: 459–464.
Sun B, Griffin BM, Ayala-del-Rio HL, Hashsham SA, Tiedje JM . (2002). Microbial dehalorespiration with 1,1,1-trichloroethane. Science 298: 1023–1025.
Tang S, Gong Y, Edwards EA . (2012). Semi-automatic in silico gap closure enabled de novo assembly of two dehalobacter genomes from metagenomic data. PLoS One 7: e52038.
Tang S, Chan WW, Fletcher KE, Seifert J, Liang X, Löffler FE et al. (2013). Functional characterization of reductive dehalogenases by using blue native polyacrylamide gel electrophoresis. Appl Environ Microbiol 79: 974–981.
Tang S, Edwards EA . (2013). Identification of Dehalobacter reductive dehalogenases that catalyse dechlorination of chloroform, 1,1,1-trichloroethane and 1,1-dichloroethane. Philos Trans R Soc Lond B Biol Sci 368: 20120318.
Tang S, Wang PH, Higgins S, Löffler F, Edwards EA . (2016). Sister Dehalobacter genomes reveal specialization in organohalide respiration and recent strain differentiation likely driven by chlorinated substrates. Front Microbiol 7: 100.
Wang S, Zhang W, Yang KL, He J . (2014). Isolation and characterization of a novel Dehalobacter species strain TCP1 that reductively dechlorinates 2,4,6-trichlorophenol. Biodegradation 25: 313–323.
Whitman WB, Wolfe RS . (1984). Purification and analysis of cobamides of Methanobacterium bryantii by high-performance liquid chromatography. Anal Biochem 137: 261–265.
Wong YK, Holland SI, Ertan H, Manefield M, Lee M . (2016). Isolation and characterization of Dehalobacter sp. strain UNSWDHB capable of chloroform and chlorinated ethane respiration. Environ Microbiol 18: 3092–3105.
Yan J, Ritalahti KM, Wagner DD, Löffler FE . (2012). Unexpected specificity of interspecies cobamide transfer from Geobacter spp. to organohalide-respiring dehalococcoides mccartyi strains. Appl Environ Microbiol 78: 6630–6636.
Yan J, Im J, Yang Y, Löffler F . (2013). Guided cobalamin biosynthesis supports Dehalococcoides mccartyi reductive dechlorination activity. Philos Trans R Soc Lond B Biol Sci 368: 20120320.
Yan J, Simsir B, Farmer AT, Bi M, Yang Y, Campagna SR et al. (2015). The corrinoid cofactor of reductive dehalogenases affects dechlorination rates and extents in organohalide-respiring Dehalococcoides mccartyi. ISME J 10: 1092–1101.
Yip SH, Matsumura I . (2013). Substrate ambiguous enzymes within the Escherichia coli proteome offer different evolutionary solutions to the same problem. Mol Biol Evol 30: 2001–2012.
Yoshida N, Ye L, Baba D, Katayama A . (2009). A novel Dehalobacter species is involved in extensive 4,5,6,7-tetrachlorophthalide dechlorination. Appl Environ Microbiol 75: 2400–2405.
Zanaroli G, Balloi A, Negroni A, Daffonchio D, Young LY, Fava F . (2010). Characterization of the microbial community from the marine sediment of the Venice lagoon capable of reductive dechlorination of coplanar polychlorinated biphenyls (PCBs). J Hazard Mater 178: 417–426.
Zelcbuch L, Lindner SN, Zegman Y, Vainberg Slutskin I, Antonovsky N, Gleizer S et al. (2016). Pyruvate formate-lyase enables efficient growth of Escherichia coli on acetate and formate. Biochem 55: 2423–2426.
Zhao S, Ding C, He J . (2015). Detoxification of 1,1,2-trichloroethane to ethene by Desulfitobacterium and identification of its functional reductase gene. PLoS One 10: e0119507.
Acknowledgements
Support was provided by the Government of Canada through Genome Canada and the Ontario Genomics Institute (2009-OGI-ABC-1405), the Government of Ontario through the ORF-GL2 program and the United States Department of Defense through the Strategic Environmental Research and Development Program (SERDP). ST received awards from the Government of Ontario through the Ontario Graduate Scholarships in Science and Technology (OGSST) and the Natural Sciences and Engineering Research Council of Canada (NSERC PGS B). We also acknowledge the BioZone Mass Spectrometry facility for UPLC-ESI-MS analyses as well as Dr Jane Howe and Mr Xu Chen (University of Toronto, Canada) and Dr Doug Holmyard (Mount Sinai Hospital, Canada) for SEM and TEM analyses. We are grateful to the generous gift of Lactobacillus delbrueckii subspecies lactis (ATCC 7831) from Agriculture Research Service (USDA).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, PH., Tang, S., Nemr, K. et al. Refined experimental annotation reveals conserved corrinoid autotrophy in chloroform-respiring Dehalobacter isolates. ISME J 11, 626–640 (2017). https://doi.org/10.1038/ismej.2016.158
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2016.158
This article is cited by
-
An interspecies malate–pyruvate shuttle reconciles redox imbalance in an anaerobic microbial community
The ISME Journal (2019)
-
Purinyl-cobamide is a native prosthetic group of reductive dehalogenases
Nature Chemical Biology (2018)
-
The organohalide-respiring bacterium Sulfurospirillum multivorans: a natural source for unusual cobamides
World Journal of Microbiology and Biotechnology (2017)