Abstract
Bacteria of the NC10 phylum link anaerobic methane oxidation to nitrite denitrification through a unique O2-producing intra-aerobic methanotrophy pathway. A niche for NC10 in the pelagic ocean has not been confirmed. We show that NC10 bacteria are present and transcriptionally active in oceanic oxygen minimum zones (OMZs) off northern Mexico and Costa Rica. NC10 16S rRNA genes were detected at all sites, peaking in abundance in the anoxic zone with elevated nitrite and methane concentrations. Phylogenetic analysis of particulate methane monooxygenase genes further confirmed the presence of NC10. rRNA and mRNA transcripts assignable to NC10 peaked within the OMZ and included genes of the putative nitrite-dependent intra-aerobic pathway, with high representation of transcripts containing the unique motif structure of the nitric oxide (NO) reductase of NC10 bacteria, hypothesized to participate in O2-producing NO dismutation. These findings confirm pelagic OMZs as a niche for NC10, suggesting a role for this group in OMZ nitrogen, methane and oxygen cycling.
Similar content being viewed by others
Main
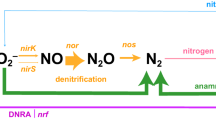
Microbes have critical roles in methane (CH4) consumption, with aerobic methane oxidation by methanotrophic bacteria and sulfate-dependent anaerobic oxidation of methane (AOM) by Archaea recognized as important sinks in the CH4 cycle (Reeburgh, 2007). Studies of freshwater habitats have further linked AOM to the reduction of oxidized nitrogen compounds, including nitrate (NO3−) and nitrite (NO2−) (Raghoebarsing et al., 2006; Haroon et al., 2013). Recently discovered NO2−-reducing bacteria of the NC10 phylum couple CH4 oxidation to N2 production through a unique NO2−-dependent anaerobic methane oxidation (n-damo) pathway (Raghoebarsing et al., 2006; Ettwig et al., 2009). Characterized in the bacterium Candidatus Methylomirabilis oxyfera from freshwater sediments (Ettwig et al., 2010), the n-damo pathway reduces NO2− to nitric oxide (NO), which is then putatively dismutated into N2 and O2 gas, with O2 serving as the oxidant for intra-aerobic methanotrophy.
The ecological significance of n-damo remains unclear, as does the environmental distribution of NC10 bacteria. Environmental evidence for NO2− or NO3−-dependent AOM stems primarily from freshwater sediments and wetlands (for example, Deutzmann et al., 2014; Norði and Thamdrup, 2014; Hu et al., 2014), while NC10 genes have been detected in diverse anoxic freshwater habitats (Shen et al., 2015), and recently in NO2−-rich sediments of the South China Sea (Chen et al., 2015). However, the potential for NC10 bacteria and n-damo in the pelagic ocean remains unknown.
Oxygen minimum zones (OMZs) are a potential niche for NO2−-dependent n-damo. In the major upwelling-driven OMZs, including the world’s largest OMZ in the Eastern Tropical North Pacific (ETNP), microbial respiration depletes [O2] to anoxia (<10 nm) (Tiano et al., 2014). The microbial reduction of NO3− to NO2− in these zones drives NO2− to micromolar concentrations (Thamdrup et al., 2012). OMZs are further characterized by elevated CH4 concentrations (Naqvi et al., 2010), and indeed represent the largest accumulation of open-ocean CH4 in the world (Sansone et al., 2001). Although sequence fragments related to NC10 bacteria (by BLAST) have been detected in OMZ meta-omic data sets (for example, Dalsgaard et al., 2014), confirmation of a pelagic OMZ niche for NC10 bacteria is lacking.
We tested the hypothesis that NO2−-replete OMZs host Methylomirabilis-like NC10 bacteria, using samples from four sites spanning a nearshore to offshore transition through the ETNP OMZ off northern Mexico and at a site in the anoxic, coastal basin of Golfo Dulce (GD), Costa Rica (Supplementary Figure S1, Supplementary Table S1). At both locations, [O2] decreased from the surface, dropping below the detection limit (20 nm) at 70–130 m depth across sites (Figure 1). Characteristic to anoxic OMZs, [NO2−] increased upon O2 depletion to a prominent maximum (4–5 μm) in the upper anoxic zone in the ETNP (120–150 m), before declining with depth. In the GD, NO2− accumulated to 0.7 μm at 100 m in the anoxic zone. Nitrite here disappeared at 140 m near the interface to a bottom layer in which hydrogen sulfide and ammonium accumulated, reaching 6.6 and 3.4 μm, respectively, at 190 m.
Water column chemistry and microbial biomass in the ETNP OMZ in May 2014 (a−d) and GD OMZ in January 2015 (e−i). ETNP stations (colors) span a nearshore (S6) to offshore (S11) gradient (Supplementary Figure S1). (a, e) Dissolved oxygen, based on standard oceanographic Clark type O2 electrodes with an inset showing higher resolution STOX sensor measurements. (f) Methane. (b, g) Nitrite. (e–i) Total (c, h) and NC10-specific (d, i) 16S rRNA gene counts, based on quantitative PCR analyses of the 0.2–1.6 μm biomass fraction. Data from the ETNP reflect discrete collections (per depth) using a CTD/rosette or a pump-profiling system during mid May 2014. Data from GD reflect discrete collections (per depth) using a hand-deployed Niskin bottle on 21 January 2015 (D1), with additional samples for 16S counts obtained on 23 January (D2). Error bars are standard deviations among replicates (duplicate and triplicate water samples from the same cast for [CH4] and [NO2-], respectively; triplicate qPCRs for 16S counts). [O2] data are provided for representative casts at each station. Challenges in calibration prevented accurate [CH4] measurements in the ETNP (see text).
Methane was present in the anoxic zone in the ETNP and GD. Challenges in methodology and calibrations prohibited us from determining exact concentrations in samples from the ENTP. Nevertheless, methane was detectable in samples from OMZ depths at ETNP sites, consistent with previous studies hypothesizing that the ETNP CH4 maximum, along an isopycnal of 26.8 kg m−3, reflects CH4 advected from coastal sediments (Sansone et al., 2001; Pack et al., 2015). The CH4 profile in the GD also suggested a sediment source, with [CH4] at trace levels above the oxycline, ~6 nm at the NO2− maximum, and ~80 nm in the sulfidic bottom water.
Quantitative PCR with NC10-specific 16S rRNA (16S) gene primers (Supplementary Table S2) detected NC10 16S genes at all OMZ sites, with maximum counts in the ETNP (25–174 copies ml−1; 0.02–0.05% of total 16S counts) at anoxic depths (~300–400 m) below the NO2− maximum (Figure 1). This distribution suggests that in addition to anoxia, ETNP NC10 bacteria are linked to CH4 rather than organic flux from above, in which case counts would be predicted to peak higher in the OMZ where organotrophic activity is highest (Babbin et al., 2014). In the GD, counts peaked at 90 m (105 copies ml−1; 0.006% of total) at the top of the NO2− and CH4-enriched zone. NC10 16S rRNA genes were undetectable or near the detection limit (6 copies ml−1) above and below the ETNP OMZ and above the GD OMZ.
Sequencing of the pmoA gene encoding particulate methane monooxygenase (pMMO), catalyzing the CH4 oxidation step in methanotrophy, further confirmed the presence of NC10. Amplification with NC10-specific primers recovered pmoA from all screened OMZ samples, representing a subset from both sites (Supplementary Table S1). Phylogenetic analysis of cloned sequences (26 from ETNP and 2 from GD) revealed four closely related phylotypes (>98.5% shared amino-acid identity) related to pmoA from NC10 environmental clones and Ca. M. oxyfera (Figure 2), with highest similarity (~98% amino-acid identity) to sequences from South China Sea sediments (Chen et al., 2015). Recovery of these sequences suggests the potential for CH4 oxidation by OMZ NC10 bacteria.
Phylogenetic evidence for the presence and transcriptional activity of NC10 in OMZs. (a) Particulate methane monooxygenase subunit A (PmoA) gene phylogeny. PmoA sequences (n=53) recovered from the ETNP and GD are highlighted in bold within the larger clade of NC10-like sequences from other studies, separated from PmoA of aerobic methanotrophic clades and an ammonia monooxygenase (AmoA) outgroup. Phylogeny was inferred based on Maximum Likelihood (ML) analysis of 88 amino acids using the Dayhoff substitution model. Bootstrap values greater than 70 are shown, along with NCBI Accession numbers for database sequences. Numbers in parentheses represent the number of unique sequences included in each collapsed node—Taxon identifiers and Accession numbers for all sequences are in Supplementary Figure S2. The scale bar represents 50 amino-acid changes per 100 amino acids. (b) Heme-copper oxidases (HCO) phylogeny following that of Ettwig et al. (2012), with putative nitric oxide dismutases (NODs) shown in green, including two full-length sequences (labeled ETNP) assembled from OMZ NC10 transcript fragments. The sequences used for assembly were those with top matches (bit score>50) to Ca. M. oxyfera via BLASTX against the NCBI-nr database. Phylogeny was inferred by ML analysis of 520 amino acids based on the Le and Gascuel_2008 model. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. (c, d) Alignment of the qNor quinol-binding (c) and catalytic (d) sites showing putative NOD (green) compared with canonical qNor (red) sequences, following that of Ettwig et al. (2012). Numbering reflects residue numbers of G. stearothermophilus. Shading highlights conserved residues in canonical qNors where amino-acid replacement has occurred in putative NODs.
Metatranscriptomes confirmed that NC10-like bacteria are transcriptionally active within OMZs. Community cDNA from five 2014 ETNP samples and three GD samples from anoxic depths was shotgun sequenced, and analyzed with five existing data sets (from Ganesh et al., 2015) from ETNP station 6 in June 2013. The relative abundance of NC10 16S rRNA transcripts increased with decreasing oxygen, peaking at the ETNP core (300 m) and at 90 m in the GD (Supplementary Tables S2 and S4), consistent with depths of maximum NC10 16S gene counts. Messenger RNA transcripts assigned to NC10 using a lowest common ancestor algorithm were detected in all data sets. NC10 mRNA transcripts varied in relative abundance and, with the exception of one sample (150 m, ETNP station 10), paralleled the distribution of NC10 rRNA, increasing with declining oxygen and peaking at ~0.1% of total mRNA transcripts in the ETNP OMZ core (Supplementary Table S2).
The OMZ NC10-like transcript pool contained genes with predicted roles in dissimilatory N transformations (Supplementary Figures S3 and S4,Supplementary Tables S5 and S6). Two norZ genes, encoding quinol-dependent NO reductases (qNor), were most abundant, together representing 51% of transcripts assignable to NC10 by the lowest common ancestor. For two samples (ETNP station 6, 300 m, 2013; station 10, 150 m, 2014), assemblies of qNor transcripts yielded full-length gene sequences that clustered phylogenetically with qNor variants (bootstrap: 100) encoded by loci DAMO_2434 and 2437 of the Ca. M. oxyfera genome (Figure 2). N2O does not appear during NO2− reduction in Ca. M. oxyfera enrichments (Ettwig et al., 2010) and it has been hypothesized that the qNor encoded by DAMO_2434 and 2437, instead of producing N2O, functions as a NO dismutase (NOD) to convert two NO molecules to N2 and O2 (Ettwig et al., 2012). Indeed, DAMO_2434 and 2437 have substitutions at key residues in both the quinol-binding and catalytic sites. The qNor sequences recovered here also lack conservation of the canonical quinol-binding motif and contain catalytic site substitutions identical to those of DAMO_2434 and 2437 (Figure 2). The identification of these candidate NODs raises the possibility of nitrite-dependent O2 production in the OMZ.
Genes of CH4 oxidation, including those encoding the pMMO enzyme, were not detected in the NC10 transcript pool identified by the lowest common ancestor. However, ETNP OMZ transcripts with top BLASTX matches to Ca. M. oxyfera pmoB were detected at low abundance in the ETNP, but not in the GD (Supplementary Table S6). pMMO-encoding transcripts of aerobic methylotrophs were not detected in the ETNP but were recovered in the upper GD OMZ (90, 100 m; data not shown), despite anoxic conditions in this zone. Low levels of NC10-assigned transcripts of CH4 oxidation may be due to low sequencing depth or sequence misclassification. Alternatively, OMZ NC10 bacteria may utilize energy substrates other than CH4.
To search for biogeochemical evidence of n-damo, we conducted anoxic 10-day incubations of OMZ water with 13CH4 and 15NO2 to measure rates of anaerobic CH4 oxidation and N2 production. CH4 oxidation rates were below the detection limit (0.6 nm d−1) at all analyzed depths in the ETNP, and at two of the three analyzed depths in the GD (100, 120 m) (Supplementary Table S7), consistent with very low published rates from the ETNP (Pack et al., 2015) and with low NC10 gene counts at both ETNP and GD sites. In contrast, CH4 oxidation was measured at 2.6±0.7 nm d−1 at 90 m in the GD and was inhibited with the addition of acetylene, a pMMO inhibitor, suggesting OMZ CH4 consumption by pMMO-catalyzed AOM. However, the recovery of MMO transcripts from putative aerobic methylotrophs at this depth suggests that methanotrophic activity was likely only to a minor extent due to NC10. N2 production rates greatly exceeded the CH4 oxidation rates in the GD, further indicating a minor contribution of NC10 to fixed nitrogen loss (Supplementary Table S7).
Together, these results identify pelagic OMZs as a niche for NC10 bacteria. Water residence time in the ETNP OMZ core is estimated at 3.9±0.8 years (DeVries et al., 2012). Given this estimate, and the observed decline in NC10 16S gene abundance with proximity to the sediment−water interface in the GD, it is unlikely that the DNA and RNA sequences detected here represent allochthonous cells advected into the system from sediment. Rather, the transcription of key diagnostic enzymes of n-damo, notably the putative NOD encoded by qNor, suggests a role for NC10 bacteria in OMZ N and O2 transformations. N2 production by NC10 would represent a route of N loss in addition to that of classical denitrification and anammox, although the latter are undoubtedly the dominant N sinks in OMZs. The recovery of NC10 pmoA genes and the vertical distribution of NC10 genes and transcripts relative to CH4 and NO2− maxima raise the possibility for NC10 CH4 consumption in OMZs, although this prediction remains to be validated. Given the observed enrichment of CH4 in OMZs, originating either from advection from sediments (Pack et al., 2015) or potentially from OMZ methanogens (Supplementary Information, Supplementary Table S8, Supplementary Figure S5), even low rates of CH4 consumption by OMZ NC10 bacteria could have an important role in the open-ocean methane budget.
References
Babbin AR, Keil RG, Devol AH, Ward BB . (2014). Organic matter stoichiometry, flux, and oxygen control nitrogen loss in the ocean. Science 344: 406–408.
Chen J, Jiang XW, Gu JD . (2015). Existence of novel phylotypes of nitrite-dependent anaerobic methane-oxidizing bacteria in surface and subsurface sediments of the South China Sea. Geomicrobiol J 32: 1–9.
Dalsgaard T, Stewart FJ, Tahmdrup B, De Brabandere L, Revsbech NP, Ulloa O et al. (2014). Oxygen at nanomolar levels reversibly suppresses process rates and gene expression in anammox and denitrification in the oxygen minimum zone off Northern Chile. Mbio 5: 10–1128.
Deutzmann JS, Stief P, Brandes J, Schink B . (2014). Anaerobic methane oxidation coupled to denitrification is the dominant methane sink in a deep lake. Proc Natl Acad Sci USA 111: 18273–18278.
DeVries T, Deutsch C, Primeau F, Chang B, Devol A . (2012). Global rates of water-column denitrification derived from nitrogen gas measurements. Nat Geosci 5: 547–550.
Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MMM et al. (2010). Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464: 543–548.
Ettwig KF, Speth DR, Reimann J, Wu ML, Jetten MS, Keltjens JT . (2012). Bacterial oxygen production in the dark. Front Microbiol 3: 273.
Ettwig KF, van Alen T, van de Pas-Schoonen KT, Jetten MSM, Strous M . (2009). Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 Phylum. Appl Environ Microbiol 75: 3656–3662.
Ganesh S, Bristow LA, Larsen M, Sarode N, Thamdrup B, Stewart FJ . (2015). Size-fraction partitioning of community gene transcription and nitrogen metabolism in a marine oxygen minimum zone. ISME J 9: 2682–2696.
Haroon MF, Hu S, Shi Y, Imelfort M, Keller J, Hugenholtz P et al. (2013). Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500: 567–570.
Hu BL, Shen LD, Lian X, Zhu Q, Liu S, Huang Q et al. (2014). Evidence for nitrite-dependent anaerobic methane oxidation as a previously overlooked microbial methane sink in wetlands. Proc Natl Acad Sci USA 111: 4495–4500.
Naqvi SWA, Bange HW, Farias L, Monteiro PMS, Scranton MI, Zhang J . (2010). Marine hypoxia/anoxia as a source of CH4 and N2O. Biogeosciences 7: 2159–2190.
Norði KA, Thamdrup B . (2014). Nitrate-dependent anaerobic methane oxidation in a freshwater sediment. Geochim Cosmochim Acta 132: 141–150.
Pack MA, Heintz MB, Reeburgh WS, Trumbore SE, Valentine DL, Xu X et al. (2015). Methane oxidation in the eastern tropical North Pacific Ocean water column. J Geophys Res Biogeosci 120: 1078–1092.
Raghoebarsing AA, Pol A, van de PasSchoonen KT, Smolders AJP, Ettwig KF, Rijpstra WIC et al. (2006). A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440: 918–921.
Reeburgh WS . (2007). Oceanic methane biogeochemistry. Chem Rev 107: 486–513.
Sansone FJ, Popp BN, Gasc A, Graham AW, Rust TM . (2001). Highly elevated methane in the eastern tropical North Pacific and associated isotopically enriched fluxes to the atmosphere. Geophys Res Lett 28: 4567–4570.
Shen LD, Wu HS, Gao ZQ . (2015). Distribution and environmental significance of nitrite-dependent anaerobic methane-oxidising bacteria in natural ecosystems. Appl Microbiol Biotechnol 99: 133–142.
Thamdrup B, Dalsgaard T, Revsbech NP . (2012). Widespread functional anoxia in the oxygen minimum zone of the Eastern South Pacific. Deep Sea Res Pt I 65: 36–45.
Tiano L, Garcia-Robledo E, Dalsgaard T, Devol AH, Ward BB, Ulloa O et al. (2014). Oxygen distribution and aerobic respiration in the north and southeastern tropical Pacific oxygen minimum zones. Deep Sea Res Pt I 94: 173–183.
Acknowledgements
We thank the crew of the R/V New Horizon for help in sample collection, Niels Peter Revsbech and Morten Larsen for oxygen concentration analysis in the GD, and Alvaro Morales for coordinating and supporting work at the GD field site. This work was supported by the National Science Foundation (1151698 to FJS), the Sloan Foundation (RC944 to FJS), the Danish National Research Foundation DNRF53, the Danish Council of Independent Research, and the European Research Council ‘Oxygen’ grant (267233; supporting LAB and BT).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Padilla, C., Bristow, L., Sarode, N. et al. NC10 bacteria in marine oxygen minimum zones. ISME J 10, 2067–2071 (2016). https://doi.org/10.1038/ismej.2015.262
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2015.262
This article is cited by
-
Water column dynamics control nitrite-dependent anaerobic methane oxidation by Candidatus “Methylomirabilis” in stratified lake basins
The ISME Journal (2023)
-
Vertical Microbial Profiling of Arabian Sea Oxygen Minimal Zone Reveals Complex Bacterial Communities and Distinct Functional Implications
Microbial Ecology (2023)
-
The marine nitrogen cycle: new developments and global change
Nature Reviews Microbiology (2022)
-
Active methane processing microbes and the disproportionate role of NC10 phylum in methane mitigation in Amazonian floodplains
Biogeochemistry (2021)
-
Pelagic denitrification and methane oxidation in oxygen-depleted waters of the Louisiana shelf
Biogeochemistry (2021)