Abstract
Haloarchaea (class Halobacteria) live in extremely halophilic conditions and evolved many unique metabolic features, which help them to adapt to their environment. The methylaspartate cycle, an anaplerotic acetate assimilation pathway recently proposed for Haloarcula marismortui, is one of these special adaptations. In this cycle, acetyl-CoA is oxidized to glyoxylate via methylaspartate as a characteristic intermediate. The following glyoxylate condensation with another molecule of acetyl-CoA yields malate, a starting substrate for anabolism. The proposal of the functioning of the cycle was based mainly on in vitro data, leaving several open questions concerning the enzymology involved and the occurrence of the cycle in halophilic archaea. Using gene deletion mutants of H. hispanica, enzyme assays and metabolite analysis, we now close these gaps by unambiguous identification of the genes encoding all characteristic enzymes of the cycle. Based on these results, we were able to perform a solid study of the distribution of the methylaspartate cycle and the alternative acetate assimilation strategy, the glyoxylate cycle, among haloarchaea. We found that both of these cycles are evenly distributed in haloarchaea. Interestingly, 83% of the species using the methylaspartate cycle possess also the genes for polyhydroxyalkanoate biosynthesis, whereas only 34% of the species with the glyoxylate cycle are capable to synthesize this storage compound. This finding suggests that the methylaspartate cycle is shaped for polyhydroxyalkanoate utilization during carbon starvation, whereas the glyoxylate cycle is probably adapted for growth on substrates metabolized via acetyl-CoA.
Similar content being viewed by others
Introduction
Microbial life exists over the whole range of salt concentration. Halophilic archaea of the class Halobacteria (haloarchaea) live in concentrated salt solutions and belong to the most important microbial habitants of saturated brine environments such as salt lakes or salterns (Oren, 2002). Their environment is photic, (micro)oxic and rich in organic nutrients, which determines them as aerobic (photo)organoheterotrophs. It is commonly anticipated that the ancestor of this highly specialized group was a strictly anaerobic, chemolithoautotrophic methanogen. Its transformation to haloarchaea, which we know today required a lateral gene transfer on a massive scale (Kennedy et al., 2001; Khomyakova et al., 2011; Nelson-Sathi et al., 2012, 2014). Therefore, haloarchaeal metabolism combines mostly laterally transferred, aerobic heterotrophic pathways from Bacteria, with some vertically inherited features of the methanogenic ancestor.
Many substrates enter the central metabolism, both its catabolic and anabolic branches, on the level of acetyl-coenzyme A (acetyl-CoA). Examples are acetate, fatty acids, alcohols, amino acids like leucine and lysine as well as the intracellular storage compound polyhydroxybutyrate. Acetyl-CoA catabolism proceeds usually through the tricarboxylic acid cycle, which provides reductant and, together with the respiratory chain, energy for biosynthetic (anabolic) reactions. Assimilation of acetyl-CoA into cellular building blocks (for example, oxaloacetate, 2-oxoglutarate, pyruvate, phosphoenolpyruvate) requires a dedicated anaplerotic pathway. Methanogenic ancestors of haloarchaea probably carboxylated acetyl-CoA in a ferredoxin-dependent pyruvate synthase reaction, as modern methanogens do (Fuchs and Stupperich, 1980; Fuchs, 2011). The functioning of this reaction in carboxylation direction needs a highly reduced ferredoxin pool that may be sustained only in anaerobic conditions (Fuchs, 2011). Aerobic haloarchaea gained two anaplerotic acetate assimilation pathways, the glyoxylate cycle and the methylaspartate cycle (Oren and Gurevich, 1995; Serrano and Bonete, 2001; Ensign, 2011; Khomyakova et al., 2011).
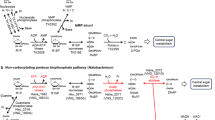
The glyoxylate cycle functioning in many bacteria, archaea and eukarya (but not in mammals) is a classical example of an anaplerotic pathway (Figure 1a; Kornberg and Krebs, 1957). Its first key enzyme, isocitrate lyase, together with enzymes of the tricarboxylic acid cycle converts acetyl-CoA into glyoxylate. The second key enzyme, malate synthase, condenses glyoxylate and a second acetyl-CoA molecule to yield malate, which can then be used as starter molecule in biosynthesis. In the methylaspartate cycle, acetyl-CoA is transformed to glutamate via reactions of the tricarboxylic acid cycle and glutamate dehydrogenase. The rearrangement of glutamate into methylaspartate and its following deamination leads to mesaconate. Mesaconate is then activated to mesaconyl-CoA, which is hydrated to β-methylmalyl-CoA, and β-methylmalyl-CoA is finally cleaved to propionyl-CoA and glyoxylate. Propionyl-CoA carboxylation leads to methylmalonyl-CoA and subsequently to succinyl-CoA, thus closing the cycle, whereas the condensation of glyoxylate with another molecule of acetyl-CoA yields the final product of the methylaspartate cycle malate (Figure 1b; Khomyakova et al., 2011). In total, two molecules of acetyl-CoA and one molecule of oxaloacetate are transformed to succinyl-CoA and malate. The emergence of both pathways in haloarchaea is probably a result of lateral gene transfer from bacteria. Haloarchaea using the glyoxylate cycle presumably acquired its key enzymes horizontally from bacteria (Serrano and Bonete, 2001). Similarly, genes for the key enzymes of the methylaspartate cycle were probably derived via lateral gene transfer from bacteria, where they originally participated in different metabolic processes (Khomyakova et al., 2011).
Anaplerotic pathways of acetyl-CoA assimilation functioning in haloarchaea. (a) The glyoxylate cycle and the tricarboxylic acid cycle (Kornberg and Krebs, 1957); (b) the methylasparate cycle (Khomyakova et al., 2011). The key reactions of the anaplerotic pathways are shown in red. Note that the methylaspartate cycle is also tightly linked to the tricarboxylic acid cycle, as both cycles share enzymes converting succinyl-CoA to 2-oxoglutarate. The names of the genes encoding key enzymes of the methylaspartate cycle are shown: mamAB, glutamate mutase (hah_1337/hah_1338); mal, methylaspartate ammonia lyase (hah_1339); mct, mesaconate CoA-transferase (hah_1336); mch, mesaconyl-CoA hydratase (hah_1340); mcl, β-methylmalyl-CoA lyase (hah_1341).
The emergence of two pathways for one and the same purpose in a phylogenetically and physiologically tight microbial group suggests the existence of an evolutionary pressure on haloarchaea selecting the species capable to assimilate compounds metabolized via acetyl-CoA. However, field studies showed that in salt lakes and salterns acetate can only be poorly utilized by haloarchaea with turnover times in the order of weeks to months (Oren, 1995). Nevertheless, haloarchaea are known to accumulate substantial amounts of polyhydroxyalkanoate, whose utilization proceeds through acetyl-CoA and hence requires an anaplerotic pathway for acetyl-CoA assimilation. Importantly, polyhydroxyalkanoates were shown to be synthesized not only in laboratory cultures but also in natural conditions (Elevi Bardavid et al., 2008). Therefore, the main function of an anaplerotic pathway in natural environments may be the assimilation of polyhydroxyalkanoate into cellular building blocks during carbon starvation rather than growth on a specific substrate. Interestingly, some haloarchaea possess genes for both cycles in the genome. This suggests the possibility that the pathways may be adapted to fulfill different functions.
The elucidation of the methylaspartate cycle was based on the combination of proteomics and enzyme activities measurements with the haloarchaeon Haloarcula marismortui (Khomyakova et al., 2011). However, activity of its key enzyme, glutamate mutase (composed of two subunits), could not be demonstrated in cells extracts of H. marismortui. This was explained by the notorious instability of this B12-dependent radical enzyme (Buckel et al., 1999; Khomyakova et al., 2011). Among enzymes of the cycle, only β-methylmalyl-CoA lyase (encoded by mcl, see Figure 2) and apparent malate synthase (malyl-CoA lyase/thioesterase) were heterologously produced and biochemically characterized (Khomyakova et al., 2011). The genes for glutamate mutase (mamAB) and methylaspartate ammonia lyase (mal) were highly similar to characterized bacterial homologs and could therefore be unambiguously identified in the genome. However, the tentative identification of succinyl-CoA:mesaconate CoA-transferase (further referred as mesaconate CoA-transferase, mct) and mesaconyl-CoA hydratase (mch) could not be experimentally confirmed. All attempts of their heterologous production resulted in the synthesis of insoluble proteins that could not be refolded in an active form (Khomyakova et al., 2011), thus questioning the identification of the genes. Furthermore, the studies of the methylaspartate cycle in H. marismortui were hampered by the lack of a genetic system for this archaeon. Therefore, we decided to switch to H. hispanica, for which a gene deletion system is available (Liu et al., 2011a) to address these important open questions.
Genomic region of Haloarcula marismortui and H. hispanica coding for the enzymes of the methylaspartate cycle (Baliga et al., 2004; Liu et al., 2011b). MamAB, glutamate mutase; Mal, methylaspartate ammonia lyase; Mct, mesaconate CoA-transferase; Mch, mesaconyl-CoA hydratase; Mcl, β-methylmalyl-CoA lyase.
H. hispanica is capable to grow on acetate as a sole carbon source and possesses a gene cluster for the methylaspartate cycle enzymes, similar to H. marismortui (Figure 2; Juez et al., 1986; Liu et al., 2011b; Ding et al., 2014). In this study, we used deletion mutants of H. hispanica to prove the participation of the key enzymes of the methylaspartate cycle in acetate assimilation. In particular, the involvement of glutamate mutase in this process could now be confirmed. Furthermore, we have confirmed the functioning of two previously unidentified genes as mesaconate CoA-transferase (mct) and mesaconyl-CoA hydratase (mch), which are crucial for the functioning of this cycle. Finally, we analyzed the distribution of the genes for the methylaspartate and glyoxylate cycles as well as genes involved in polyhydroxyalkanoate biosynthesis in sequenced haloarchaeal genomes. Based on this analysis, we hypothesize that the methylaspartate cycle represents a metabolic adaptation to the utilization of polyhydroxybutyrate during carbon starvation.
Materials and methods
Microbial strains and culture conditions
The strains used in this study are listed in Supplementary Table 1. H. hispanica strain ATCC 33960 was grown under aerobic conditions at 37 °C in a 10-l flask using an air pump or in shaken 1-l flasks on a chemically defined medium (Kauri et al., 1990) with acetate (0.2%, w/v), pyruvate (0.2%) or acetate (0.2%) plus pyruvate (0.2%) as carbon sources. For the cultivation of the uracil auxotrophic (pyrF-deleted) strain DF60 (Liu et al., 2011a) and its derivatives, uracil (stock solution 50 mg ml−1, dissolved in DMSO) was added at a concentration of 50 mg l−1.
Materials
Chemicals were obtained from Fluka (Neu-Ulm, Germany), Sigma-Aldrich (Deisenhofen, Germany), Merck (Darmstadt, Germany), Serva (Heidelberg, Germany) or Roth (Karlsruhe, Germany). Biochemicals were from Roche Diagnostics (Mannheim, Germany), AppliChem (Darmstadt, Germany) or Gerbu (Craiberg, Germany). Materials for cloning and expression were purchased from MBI Fermentas (St Leon-Rot, Germany), New England Biolabs (Frankfurt, Germany), Novagen (Schwalbach, Germany), Genaxxon Bioscience GmbH (Biberach, Germany), MWG Biotech AG (Ebersberg, Germany), Biomers (Ulm, Germany) or Qiagen (Hilden, Germany). Primers were synthesized by Thermo Fisher Scientific-Invitrogen (Beijing, China).
Syntheses
Acetyl-CoA, propionyl-CoA and succinyl-CoA were synthesized from their anhydrides by the method explained by Simon and Shemin (1953). β-Methylmalyl-CoA was synthesized enzymatically with recombinant (S)-malyl-CoA/β-methylmalyl-CoA/(S)-citramalyl-CoA lyase from Chloroflexus aurantiacus (Zarzycki et al., 2009), as described previously (Sasikaran et al., 2014). A mixture of mesaconyl-C1-CoA and mesaconyl-C4-CoA was synthesized chemically from the free acid by the mixed anhydride method of Stadtman (1957). From this mixture, mesaconyl-C1-CoA was purified using high-performance liquid chromatography (Zarzycki et al., 2009). The dry powders of the CoA-esters were stored at −20 °C.
Mutant construction and verification
The strains, plasmids and primers used for mutant construction and verification are listed in Supplementary Tables 1 and 2. Mutant construction was performed using the pop-in/pop-out method described previously (Liu et al., 2011a). The gene deletion mutants were selected by PCR verification and confirmed by sequence analysis. To complement the gene deletion mutants, the putative promoter sequence of hah_1336 (109-bp-upstream region of hah_1336) and the hah_1336, hah_1338, hah_1339 or hah_1340 genes were linked and inserted into the plasmid pWL502, resulting in the plasmids pWLHah1336, pWLHah1338, pWLHah1339 and pWLHah1340, which were transformed into the respective mutant strains. The transformation of H. hispanica was performed using the polyethylene glycol-mediated method (Cline et al., 1989).
Preparation of cell extracts
Cell extracts were prepared under oxic conditions using a mixer-mill (MM200, Retsch, Haare, Germany). Cells (100–150 mg) were suspended in 0.5 ml of 20 mM 3-(N-morpholino)propanesulfonic acid (MOPS)/KOH, pH 6.9, 0.1 mg ml−1 DNase I and 0.5 mM dithiothreitol in 1.5 ml Eppendorf vials. Then, 1 g of glass beads (diameter 0.1–0.25 mm) was added to the suspension, and the cells were treated in the mixer-mill for 10 min at 30 Hz. This was followed by a centrifugation step (14 000 g, 4 °C, 10 min), and the supernatant (cell extract) was used for enzyme assays.
Enzyme assays
Spectrophotometric enzyme assays (0.5 ml assay mixture) were performed aerobically in 0.5 ml cuvettes at 37 °C. Reactions involving NAD(P)H were measured at 365 nm (ɛNADH=3.4 mM−1 cm−1, ɛNADPH=3.5 mM−1 cm−1; Bergmeyer, 1975). Reactions with phenylhydrazine were measured at 324 nm (ɛglyoxylate phenylhydrazone=17 mM−1 cm−1; Herter et al., 2001). Reactions with 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) were measured at 412 nm (ɛDTNB-CoA=14.2 mM−1 cm−1; Riddles et al., 1983).
Citrate synthase and malate synthase activities were detected spectrophotometrically as oxaloacetate- and glyoxylate-dependent release of CoA from acetyl-CoA with DTNB, isocitrate dehydrogenase activity as isocitrate-dependent reduction of NAD(P)+, isocitrate lyase activity by monitoring glyoxylate formation from isocitrate with phenylhydrazine, crotonyl-CoA carboxylase/reductase and malonyl-CoA reductase activities as crotonyl-CoA- and malonyl-CoA-dependent NAD(P)H oxidation, and glutamate dehydrogenase as 2-oxoglutarate-dependent NAD(P)H oxidation, as described previously (Khomyakova et al., 2011). Methylaspartate ammonia lyase and glutamate mutase were measured with high-performance liquid chromatography according to a previously published procedure (Khomyakova et al., 2011).
Mesaconate:succinyl-CoA CoA-transferase activity was measured by ultra-performance liquid chromatography (UPLC) as succinyl-CoA- and mesaconate-dependent formation of mesaconyl-CoA (and the products of its further conversion, β-methylmalyl-CoA and propionyl-CoA). The reaction mixture contained 100 mM Tris/HCl (pH 7.8), 3 M KCl, 5 mM MgCl2, 3.5 mM phenylhydrazine, succinyl-CoA (1 mM) and cell extract. The reaction was started by the addition of 10 mM mesaconate. After appropriate time intervals, 25 μl of the assay mixture was transferred to ice and stopped by addition of 5 μl of 1 M HCl. Protein was removed by centrifugation, and the samples were analyzed by reverse-phase (RP) C18 UPLC.
Mesaconyl-CoA hydratase was measured either in forward or reverse direction by the formation of β-methylmalyl-CoA (and propionyl-CoA) from mesaconyl-C1-CoA or mesaconyl-C1-CoA from β-methylmalyl-CoA, respectively. For the measurement in the forward direction, the reaction mixture contained 100 mM MOPS/KOH (pH 7.0), 3 M KCl, 5 mM MgCl2, 3.5 mM phenylhydrazine, 0.5 mM mesaconyl-C1-CoA and cell extract. For the measurement in the reverse direction, phenylhydrazine was omitted from the mixture, and mesaconyl-C1-CoA was replaced by β-methylmalyl-CoA (0.5 mM). The reaction was started by the addition of CoA-ester and stopped as described above. The products were analyzed by RP-C18 UPLC.
β-Methylmalyl-CoA lyase was measured either in forward or reverse direction by the formation of propionyl-CoA from β-methylmalyl-CoA or β-methylmalyl-CoA from propionyl-CoA and glyoxylate, respectively. For the measurement in the forward direction, the reaction mixture contained 100 mM MOPS/KOH (pH 7.0), 3 M KCl, 5 mM MgCl2, 3.5 mM phenylhydrazine, 0.5 mM β-methylmalyl-CoA and cell extract. For the measurement in the reverse direction, the reaction mixture contained 100 mM MOPS/KOH (pH 7.0), 3 M KCl, 5 mM MgCl2, 1 mM propionyl-CoA, 10 mM glyoxylate and cell extract. The reaction was started by the addition of CoA-ester and stopped after appropriate time intervals, as described above. The products were analyzed by RP-C18 UPLC.
Propionyl-CoA carboxylase was measured as propionyl-CoA-dependent formation of methylmalonyl-CoA in the reaction mixture containing 100 mM Tris/HCl (pH 7.8), 5 mM dithiothreitol, 3 M KCl, 5 mM MgCl2, 3 mM ATP, 15 mM NaHCO3, 1 mM propionyl-CoA and cell extract. The reaction was stopped after appropriate time intervals, as described above, and the products were analyzed by RP-C18 UPLC.
Mesaconate and succinyl-CoA conversion to propionyl-CoA and glyoxylate
The reaction mixture contained 100 mM MOPS/KOH (pH 7.0), 3 M KCl, 5 mM MgCl2, 3.5 mM phenylhydrazine, 2 mM succinyl-CoA, 10 mM mesaconate and cell extract. In a control, mesaconate was omitted. The reaction was stopped at appropriate time points by transferring 25 μl of the mixture to 5 μl 1 M HCl, protein was removed by centrifugation and samples were analyzed by RP-C18 UPLC.
Determination of glutamate and mesaconate concentrations in the cytoplasm of H. hispanica
Glutamate concentration was determined, as described previously (Khomyakova et al., 2011). For the determination of intracellular mesaconate concentration, 100 mg of the cells were dissolved in 400 μl of distilled water with traces of DNase I and vortexed for 20 min. After 1 h incubation at room temperature, cell lysates were centrifuged 15 min at 14 000 g (4 °C). The mesaconate concentration in supernatant was determined using the Aminex HPX-87H ion exclusion column high-performance liquid chromatography, and the obtained values were correlated with the volume of cytoplasm based on the assumption that 0.5 g protein in the sample corresponds to 1 g of dry cell weight and to 2 ml of free water in the cytoplasm (Khomyakova et al., 2011).
Database search and phylogenetic analysis
Query sequences were obtained from the National Center for Biotechnology Information database. The BLAST searches were performed via National Center for Biotechnology Information BLAST server (http://www.ncbi.nlm.nih.gov/BLAST/; Altschul et al., 1990). The amino-acid sequences were aligned with sequences from GenBank using CLUSTALW (Thompson et al., 1994) implemented within BioEdit software (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). The phylogenetic tree was reconstructed using a maximum likelihood algorithm (Felsenstein, 1981) in the MEGA6 program (Tamura et al., 2013). 1000 bootstrap replication were conducted to evaluate the reliabilities of the reconstructed trees. The GenBank accession numbers for the RNA polymerase subunit B’ and PhaC protein sequences are listed in Supplementary Tables 3 and 4.
Other methods
CoA and CoA-esters were identified and quantified by UPLC using a reversed phase C18 column (BEH C18, 1.7 μm, 2.1 × 100 mm column, Waters, Eschborn, Germany), as described in Sasikaran et al. (2014). In vivo specific carbon fixation rate of H. hispanica was calculated from the generation time of the culture, as described previously (Sasikaran et al., 2014). Protein was measured according to the Bradford method (Bradford, 1976), using bovine serum albumin as a standard. DNA sequence determination was performed by GATC Biotech (Constance, Germany) and Thermo Fisher Scientific-Invitrogen (Beijing, China).
Results
Acetate assimilation in wild-type H. hispanica cells
H. hispanica grew on media with acetate as a sole carbon source with a generation time of 24 h, which corresponds to a specific growth rate of 0.029 h−1 and a specific carbon assimilation rate of 37.5 nmol min−1 mg−1 protein. Because one C4-compound is synthesized from two acetyl-CoA molecules in the methylaspartate cycle (Figure 1), the minimal in vivo specific activity of enzymes of the cycle is 9.4 nmol min−1 mg−1 protein. As a part of cellular carbon is synthesized directly from acetyl-CoA and is not generated in an anaplerotic pathway, the actual minimal in vivo specific activity of enzymes of the cycle is even slightly lower.
The results of enzyme assays confirmed the absence of the key enzymes of the glyoxylate cycle (isocitrate lyase), of the ethylmalonyl-CoA pathway (crotonyl-CoA carboxylase/reductase, Erb et al., 2007), and of the 3-hydroxypropionate bi-cycle (malonyl-CoA reductase, Zarzycki et al., 2009) in H. hispanica (Table 1). On the other hand, key enzymes of the methylaspartate cycle were found to be active in acetate-grown H. hispanica cells. The only exception was glutamate mutase, which is known to be highly unstable (Buckel et al., 1999) and was not detected in H. marismortui as well (Khomyakova et al., 2011). The measured activities were sufficient to explain the observed growth rate. Activities of the enzymes were not detected in pyruvate-grown cells, thus confirming their involvement in acetate assimilation (Table 1). Furthermore, cell extracts of acetate-grown H. hispanica catalyzed the conversion of mesaconate into propionyl-CoA and glyoxylate at a specific rate of 7.7 nmol min−1 mg−1 protein, when succinyl-CoA was added as a CoA donor (Figure 3). In this conversion, the intermediary formation of the postulated intermediates mesaconyl-CoA and β-methylmalyl-CoA was observed. Therefore, not only the activities of the single enzymes responsible for mesaconate conversion to propionyl-CoA and glyoxylate but also the whole reaction sequence could be demonstrated in vitro.
Conversion of mesaconate to propionyl-CoA and glyoxylate by cell extracts of acetate-grown H. hispanica. The experiment was started by the addition of succinyl-CoA, and the samples after 0 (a), 5 (b) and 10 min (c) of incubation were analyzed by reversed phase C18 UPLC to follow CoA-esters (at 260 nm) and glyoxylate phenylhydrazine (at 324 nm). (d) Control without mesaconate after 10 min of incubation. Increase of the CoA peak in the control is due to succinyl-CoA hydrolysis. Abs260, absorption at 260 nm; Abs324, absorption at 324 nm; Gl-Ph, glyoxylate phenylhydrazone.
The functioning of the methylaspartate cycle requires a high intracellular concentration of glutamate (Khomyakova et al., 2011). The reasons for that are the unfavorable equilibrium of glutamate mutase reaction (that is, on the side of the substrate glutamate) and the low affinity of methylaspartate ammonia lyase for methylaspartate. Indeed, the cytoplasmic glutamate concentration in H. hispanica was 153±66 and 155±88 mM for acetate- and pyruvate-grown cells, respectively. Therefore, the methylaspartate cycle appears to be a glutamate overflow mechanism: it functions only at glutamate concentrations, which exceed a certain threshold signalizing the availability of nitrogen and energy to perform anabolic reactions (see Discussion).
Experiments with H. hispanica gene deletion mutants



To study genes involved in the methylaspartate cycle, we made pop-in/pop-out deletions of four genes (Figure 2): one of the two glutamate mutase genes mamB (Δhah_1338 strain), methylaspartate ammonia lyase gene mal (Δhah_1339 strain), putative mesaconate CoA-transferase gene mct (Δhah_1336 strain) and mesaconyl-CoA hydratase gene mch (Δhah_1340 strain). All mutants grew with pyruvate and pyruvate plus acetate. However, none of these strains was capable to grow in the medium containing acetate as the only carbon source (Figure 4). Complementation of these mutant strains with the corresponding genes under control of the native promoter of hah_1336 gene (its 109-bp-upstream region) using a plasmid pWL502 restored the growth on acetate (Supplementary Figure 1), thus confirming the absence of polar effect on the expression of the adjacent genes. These results showed that these four characteristic enzymes are indeed vital for the functioning of the postulated methylaspartate cycle.
Growth of Haloarcula hispanica wild-type strain (a), ΔmamB (Δhah_1338) (b), Δmal (Δhah_1339) (c), Δmct (Δhah_1336) (d), Δmch (Δhah_1340) (e) on the medium with acetate (), pyruvate () or acetate and pyruvate (). The experiment was done in triplicate, the error bars represent the standard deviations. Note that the mutants did not grow on the medium with acetate alone even after 21 days of incubation. The Δmct mutant grows slowly on the medium with pyruvate compared with the wild-type strain. The reason for that is unknown.
To confirm the function of the disrupted genes, we decided to measure the enzymes of the methylaspartate cycle in the mutants. However, the wild-type strain synthesized enzymes of the cycle only during growth on acetate in the absence of a second substrate (Table 1), and the mutants did not grow on the medium with acetate as a single-carbon source. To overcome these obstacles, we grew the cells on pyruvate, and then harvested, washed and incubated them in acetate medium. Using this approach, we were able to detect activities of the methylaspartate cycle enzymes in both wild-type and mutant cells (Table 2). The activity of the methylaspartate ammonia lyase was not detected in the corresponding mutant (Δmal), thus supporting the function of the respective gene product. The activities of other enzymes of the cycle were detected in this mutant, implying the absence of a polar effect of the mutation on the other genes. A similar phenotype was observed for the Δhah_1336 and Δhah_1340 mutants. In these mutant strains, only the activities of mesaconate CoA-transferase (Δhah_1336) and mesaconyl-CoA hydratase (Δhah_1340) were missing (Table 2). This allows the unambiguous identification of these genes as a mesaconate CoA-transferase (hah_1336, mct) and a mesaconyl-CoA hydratase (hah_1340, mch) of the methylaspartate cycle.
Further evidence for the operation of the methylaspartate cycle in H. hispanica was provided by the analysis of the metabolite concentrations in pyruvate-grown cells incubated in acetate medium. Mesaconate, a specific intermediate of the methylaspartate cycle, was detected in the wild-type (DF60) cells, whereas this compound was not detected in the glutamate mutase (ΔmamB) and methylaspartate ammonia lyase (Δmal) mutants (Table 3). These data confirm that H. hispanica synthesizes mesaconate from glutamate via methylaspartate, even though the activity of the key enzyme of this conversion, glutamate mutase, was not be measured in cell extracts, probably due to its instability. On the contrary, the mesaconate CoA-transferase and mesaconyl-CoA hydratase mutants accumulated large amounts of mesaconate in the cells (Table 3). This suggests that further metabolism of mesaconate is blocked in Δmct (Δhah_1336) and Δmch (Δhah_1340) strains, additionally proving the identification of Hah_1336 and Hah_1340. As the methylaspartate cycle is the only known haloarchaeal metabolic pathway with mesaconate as an intermediate, this observation strongly supports the operation of the cycle in vivo. Glutamate levels were lower in all mutants compared with the wild-type strain (Table 3), reflecting the requirement of a functional anaplerotic pathway to synthesize glutamate and to maintain its pool (and the pools of other cellular building blocks).
Distribution of the genes for the methylaspartate and glyoxylate cycles in haloarchaea
In addition to verifying experimentally the functioning of the methylaspartate cycle in haloarchaea, we were also able to prove experimentally the identity of all genes for key enzymes of the cycle. Using this information, we analyzed the distribution of the genes for the methylaspartate and glyoxylate cycles in the genomes of 102 sequenced species. The full set of the genes for the methylaspartate cycle was found in the genomes of 42 species of haloarchaea (41%), whereas isocitrate lyase and malate synthase genes were found in 56 haloarchaeal genomes (55%; Figure 5). Three haloarchaea (Natrialba magadii, N. chahannaoensis and Natronolimnobius innermongolicus) possess genes for both pathways. Interestingly, the distribution of these two acetate assimilation pathways fits well to the current taxonomy of haloarchaea (class Halobacteria): the methylaspartate cycle is used by the representatives of the order Natrialbales, whereas the glyoxylate cycle is present in Haloferacales (Figure 5). Although most of Halobacteriales possess the glyoxylate cycle, the representatives of Haloarcula/Halomicrobium group use the methylaspartate cycle for acetyl-CoA assimilation.
Phylogeny of the class Halobacteria based on the analysis of RNA polymerase subunit B’ (RpoB’) protein sequences. The tree shows the pattern of characteristic enzymes involved in haloarchaeal carbon metabolism (according to the data of the genome analysis), that is, the presence of the characteristic enzymes of the glyoxylate cycle (isocitrate lyase) and of the methylaspartate cycle (glutamate mutase, methylaspartate ammonia lyase, succinyl-CoA:mesaconate CoA-transferase, mesaconyl-CoA hydratase and β-methylmalyl-CoA lyase) as well as of the homologs of haloarchaeal polyhydroxyalkanoate synthase (Han et al., 2010). Growth on the substrates metabolized via acetyl-CoA (acetate, fatty acids, tween, leucine and/or lysine) is according to descriptions of the corresponding strains/species. Numbers at nodes indicate the percentage bootstrap values for the clade of this group in 1000 replications. The branches with the values below 50% are drawn as unresolved. The accession numbers of the sequences used for the construction of the tree are listed in Supplementary Table 3.
The presence of the methylaspartate cycle often coincides with the capability of haloarchaea to synthesize polyhydroxyalkanoates. 83% of the species using the methylaspartate cycle possess also polyhydroxyalkanoate synthase (Han et al., 2010) and are thus probably capable to synthesize polyhydroxyalkanoates. In contrast, only 34% of the species with the glyoxylate cycle have the genetic potential to synthesize this storage material (Figures 5 and 6). These data suggest a correlation between the capability to accumulate polyhydroxyalkanoates and the choice of the acetate assimilation pathway. Note that the phylogenetic tree of the polyhydroxyalkanoate synthase PhaC (Supplementary Figure 2) correlates well with the RNA polymerase subunit B’ tree (Figure 5), thus implying that the ancestors of all three orders possessed polyhydroxyalkanoates synthase genes and that these genes were transferred mainly vertically in haloarchaea.
Discussion
This work evidences the functioning of the methylaspartate cycle in Haloarcula using enzyme assays and studies of deletion mutants of key genes of the pathway. We have shown the presence of the enzymes of the cycle in the wild-type H. hispanica cells, the failure of the mutants to grow with acetate as the sole carbon source, and the accumulation of the specific intermediate of the cycle, mesaconate, if the reactions after mesaconate formation are blocked. The distinct growth phenotype of the glutamate mutase mutant clearly shows the involvement of this enzyme in acetate assimilation, even though we could not measure the activity of this enzyme in haloarchaea. Furthermore, by incubating the mutants in the presence of acetate and measuring the activities of the enzymes of the cycle, we could confirm the identification of the genes for mesaconate CoA-transferase and mesaconyl-CoA hydratase. The prior preliminary identification of these genes did not have an experimental support and was based only on their co-localization in the genome with the genes for glutamate mutase, methylaspartate ammonia lyase and β-methylmalyl-CoA lyase of the cycle and relied on their putative general functions (CoA-transferase, enoyl-CoA hydratase).
The analysis of the genomes of the sequenced haloarchaea for the genes of the methylaspartate cycle shows that this is a widespread and successful strategy of acetyl-CoA assimilation in this archaeal group (Figure 5). Haloarchaea have recently been separated into three orders, namely Halobacteriales, Haloferacales and Natrialbales (Gupta et al., 2014). The methylaspartate cycle is distributed among Natrialbales and some Halobacteriales, whereas most of the Haloferacales and Halobacteriales possess the glyoxylate cycle (Figure 5). The genome of one species of Haloferacales (Halogranum salarium) encodes the methylaspartate cycle genes, and three species of Natrialbales possess the genes for the glyoxylate cycle in addition to those of the methylaspartate cycle. This may be explained by the ability of haloarchaea to frequently exchange DNA between species, thus disrupting co-evolution of the genes and complicating the evolutionary history of this group (Williams et al., 2012; DeMaere et al., 2013). Interestingly, the representatives of the genus Halobacterium possess only glutamate mutase and methylaspartate ammonia lyase genes, although the genes for other key enzymes of the methylaspartate cycle are missing. They probably use the corresponding enzymes for the utilization of glutamate (Falb et al., 2008).
The genome analysis suggests that the usage of the methylaspartate cycle often correlates with the capability to synthesize polyhydroxyalkanoates, whereas the haloarchaea using the glyoxylate cycle are often incapable to synthesize this storage material (Figure 6). How could this be explained? What are the differences between the glyoxylate and methylaspartate cycles in this respect?
Bacteria using the glyoxylate cycle for acetyl-CoA assimilation should find a way to separate the fluxes between the glyoxylate cycle and the tricarboxylic acid cycle. Isocitrate, the intermediate at the branch point of these two cycles, can react either with isocitrate dehydrogenase leading into the energy-producing steps of the tricarboxylic acid cycle, or with isocitrate lyase (Figure 1a). This means that isocitrate lyase and isocitrate dehydrogenase directly compete for isocitrate. The kinetic parameters of isocitrate dehydrogenase are normally better than those of isocitrate lyase: for example, the Haloferax volcanii enzymes have the Km value to isocitrate of 0.13 and 1.16 mM, respectively (Camacho et al., 1995; Serrano et al., 1998). Therefore, the functioning of isocitrate lyase in the presence of isocitrate dehydrogenase requires a large pool of isocitrate, overproduction of isocitrate lyase, or complicated regulation mechanisms controlling isocitrate dehydrogenase activity like, for example, phosphorylation in enterobacteria (El-Mansi et al., 1994; Cozzone and El-Mansi, 2005). Alternatives are synthesis of different isoforms of isocitrate dehydrogenase with different kinetic constants, depending on conditions (Reeves et al., 1983, 1986; Banerjee et al., 2005), or eukaryotic-specific mechanism of spatial separation of the glyoxylate and tricarboxylic acid cycles, like it happens in glyoxysomes of fungi and plants (Graham, 2008; Strijbis and Distel, 2010). However, these mechanisms are not functional in haloarchaea, and accumulation of large amount of isocitrate or isocitrate lyase is possible only in actively growing culture. Therefore, the glyoxylate cycle may not represent an appropriate pathway for assimilation of polyhydroxyalkanoates in carbon starvation conditions.
The methylaspartate cycle branches off the tricarboxylic acid cycle on the level of 2-oxoglutarate (Figure 1b). 2-Oxoglutarate is converted to glutamate, which is the dominant carrier of amino groups in metabolism. The rate of this conversion depends on ammonium availability and energy supply (Leigh and Dodsworth, 2007), thus turning on the methylaspartate cycle under conditions when anabolism is desirable and carbon needs to be supplied for biosynthesis. It allows the separation of the flows of the methylaspartate and tricarboxylic acid cycles, thus preventing the competition between these two cycles for intermediates. Therefore, the methylaspartate cycle is well adapted for the assimilation of acetyl-CoA produced from the internal carbon storage during carbon starvation (Figure 1). This feature is especially important for haloarchaea living under conditions of frequent starvation periods and intermittently occurring blooms when the biomass (and storage compounds) are produced (Oren, 2002). Therefore, we hypothesize that the methylaspartate cycle is a preferential strategy for haloarchaea that should rely on assimilation of storage compounds in their natural habitats. However, this hypothesis requires further experimental support.
Glutamate concentration in actively growing Haloarcula cells was shown to be as high as 26 mM (in H. marismortui, Khomyakova et al., 2011) and 150 mM (in H. hispanica, this work), whereas acetate-grown Haloferax volcanii possesses only 6 mM glutamate in its cytoplasm (Khomyakova et al., 2011). Although accumulation of high concentrations of KCl is the main strategy used by haloarchaea to balance their cytoplasm osmotically with their medium (Oren, 2002), high intracellular glutamate concentration may be advantageous for a halophilic organism. Indeed, glutamate is a well-known osmolyte used by many halophiles (Oren, 2002) and is the precursor for the intracellular antioxidant γ-glutamylcystein used by haloarchaea (Malki et al., 2009).
Haloarchaea have been shown to synthesize a co-polymer, poly(3-hydroxybutyrate-co-3-hydroxyvalerate), rather than a homopolymer polyhydroxybutyrate (Han et al., 2009; Quillaguamán et al., 2010). Degradation of this co-polymer leads to the formation of acetyl-CoA and propionyl-CoA, and the methylaspartate cycle allows the co-assimilation of these two compounds. This is an important feature of the cycle, as propionyl-CoA inhibits several enzymes of the central metabolism (Brock and Buckel, 2004; Hou et al., 2015), and impaired propionyl-CoA metabolism affects growth of haloarchaea (Hou et al., 2015). Furthermore, acetate and propionate are often found in the same environments, and their simultaneous utilization through the methylaspartate cycle may be advantageous.
Interestingly, the ethylmalonyl-CoA pathway, another alternative to the glyoxylate cycle functioning in various proteobacteria and streptomycetes (Erb et al., 2007, 2009), involves formation of acetoacetyl-CoA, that is also an intermediate of polyhydroxybutyrate biosynthesis and degradation (Dawes, 1988). Like the methylaspartate cycle, the ethylmalonyl-CoA pathway involves the intermediates mesaconyl-CoA and methylmalyl-CoA and both pathways result in the formation and assimilation of propionyl-CoA. Apparently, the ethylmalonyl-CoA pathway works in concert with the polydroxyalkanoate metabolism and is therefore well-suited for the assimilation of this storage material. The suitability of an anaplerotic pathway for the usage of polyhydroxyalkanoates may be one of the driving forces in the evolution of alternative acetyl-CoA assimilation strategies.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ . (1990). Basic local alignment search tool. J Mol Biol 215: 403–410.
Baliga NS, Bonneau R, Facciotti MT, Pan M, Glusman G, Deutsch EW et al. (2004). Genome sequence of Haloarcula marismortui: a halophilic archaeon from the Dead Sea. Genome Res 14: 2221–2234.
Banerjee S, Nandyala A, Podili R, Katoch VM, Hasnain SE . (2005). Comparison of Mycobacterium tuberculosis isocitrate dehydrogenases (ICD-1 and ICD-2) reveals differences in coenzyme affinity, oligomeric state, pH tolerance and phylogenetic affiliation. BMC Biochem 6: 20.
Bergmeyer HU . (1975). Neue Werte für die molaren Extinktions-Koeffizienten von NADH und NADPH zum Gebrauch im Routine-Laboratorium. Z Klin Chem Klin Biochem 13: 507–508.
Bradford MM . (1976). A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254.
Brock M, Buckel W . (2004). On the mechanism of action of the antifungal agent propionate. Eur J Biochem 271: 3227–3241.
Buckel W, Broker G, Bothe H, Pierik AJ, Golding BT . (1999) Glutamate mutase and 2-methyleneglutarate mutase. In: Chemistry and Biochemistry of B12 . Banerjee R (ed). Wiley: : New York, pp 757–781.
Camacho ML, Brown RA, Bonete MJ, Danson MJ, Hough DW . (1995). Isocitrate dehydrogenases from Haloferax volcanii and Sulfolobus solfataricus: enzyme purification, characterisation and N-terminal sequence. FEMS Microbiol Lett 134: 85–90.
Cline SW, Lam WL, Charlebois RL, Schalkwyk LC, Doolittle WF . (1989). Transformation methods for halophilic archaebacteria. Can J Microbiol 35: 148–152.
Cozzone AJ, El-Mansi M . (2005). Control of isocitrate dehydrogenase catalytic activity by protein phosphorylation in Escherichia coli . J Mol Microbiol Biotechnol 9: 132–146.
Dawes EA . (1988). Polyhydroxybutyrate: an intriguing biopolymer. Bioscience Rep 8: 537–547.
DeMaere MZ, Williams TJ, Allen MA, Brown MV, Gibson JA, Rich J et al. (2013). High level of intergenera gene exchange shapes the evolution of haloarchaea in an isolated Antarctic lake. Proc Natl Acad Sci USA 110: 16939–16944.
Ding JY, Chiang PW, Hong MJ, Dyall-Smith M, Tang SL . (2014). Complete genome sequence of the extremely halophilic archaeon Haloarcula hispanica strain N601. Genome Announc 2: e00178–14.
Elevi Bardavid R, Khristo P, Oren A . (2008). Interrelationships between Dunaliella and halophilic prokaryotes in saltern crystallizer ponds. Extremophiles 12: 5–14.
El-Mansi EM, Dawson GC, Bryce CF . (1994). Steady-state modelling of metabolic flux between the tricarboxylic acid cycle and the glyoxylate bypass in Escherichia coli . Comput Appl Biosci 10: 295–299.
Ensign SA . (2011). Another microbial pathway for acetate assimilation. Science 331: 294–295.
Erb TJ, Berg IA, Brecht V, Müller M, Fuchs G, Alber BE . (2007). Synthesis of C5-dicarboxylic acids from C2-units involving crotonyl-CoA carboxylase/reductase: the ethylmalonyl-CoA pathway. Proc Natl Acad Sci USA 104: 10631–10636.
Erb TJ, Fuchs G, Alber BE . (2009). (2S)-Methylsuccinyl-CoA dehydrogenase closes the ethylmalonyl-CoA pathway for acetyl-CoA assimilation. Mol Microbiol 73: 992–1008.
Falb M, Müller K, Königsmaier L, Oberwinkler T, Horn P, von Gronau S et al. (2008). Metabolism of halophilic archaea. Extremophiles 12: 177–196.
Felsenstein J . (1981). Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17: 368–376.
Fuchs G . (2011). Alternative pathways of carbon dioxide fixation: insights into the early evolution of life? Annu Rev Microbiol 65: 631–658.
Fuchs G, Stupperich E . (1980). Acetyl CoA, a central intermediate of autotrophic CO2 fixation in Methanobacterium thermoautotrophicum . Arch Microbiol 127: 267–272.
Graham IA . (2008). Seed storage oil mobilization. Annu Rev Plant Biol 59: 115–142.
Gupta RS, Naushad S, Baker S . (2014). Phylogenomic analyses and molecular signatures for the class Halobacteria and its two major clades: a proposal for division of the class Halobacteria into an emended order Halobacteriales and two new orders, Haloferacales ord. nov. and Natrialbales ord. nov., containing the novel families Haloferacacaeae fam. Nov. and Natrialbaceae fam. nov. Int J Syst Evol Microbiol. 65: 1050–1069.
Han J, Lu Q, Zhou L, Liu H, Xiang H . (2009). Identification of the polyhydroxyalkanoate (PHA)-specific acetoacetyl coenzyme A reductase among multiple FabG paralogs in Haloarcula hispanica and reconstruction of the PHA biosynthetic pathway in Haloferax volcanii . Appl Environ Microbiol 75: 6168–6175.
Han J, Hou J, Liu H, Cai S, Feng B, Zhou J, Xiang H . (2010). Wide distribution among halophilic archaea of a novel polyhydroxyalkanoate synthase subtype with homology to bacterial type III synthases. Appl Environ Microbiol 76: 7811–7819.
Herter S, Farfsing J, Gad’on N, Rieder C, Eisenreich W, Bacher A, Fuchs G . (2001). Autotrophic CO2 fixation by Chloroflexus aurantiacus: study of glyoxylate formation and assimilation via the 3-hydroxypropionate cycle. J Bacteriol 183: 4305–4316.
Hou J, Xiang H, Han J . (2015). Propionyl coenzyme A (propionyl-CoA) carboxylase in Haloferax mediterranei: Indispensability for propionyl-CoA assimilation and impacts on global metabolism. Appl Environ Microbiol 81: 794–804.
Juez G, Rodriguez-Valera F, Ventosa A, Kushner DJ . (1986). Haloarcula hispanica spec. nov. and Haloferax gibbonsii spec. nov., two new species of extremely halophilic archaebacteria. Syst Appl Microbiol 8: 75–79.
Kauri T, Wallace R, Kushner DJ . (1990). Nutrition of the halophilic archaebacterium. Haloferax volcanii. Syst Appl Microbiol 13: 14–18.
Kennedy SP, Ng WV, Salzberg SL, Hood L, DasSarma S . (2001). Understanding the adaptation of Halobacterium species NRC-1 to its extreme environment through computational analysis of its genome sequence. Genome Res 11: 1641–1650.
Khomyakova M, Bükmez Ö, Thomas LK, Erb TJ, Berg IA . (2011). A methylaspartate cycle in haloarchaea. Science 331: 334–337.
Kornberg HL, Krebs HA . (1957). Synthesis of cell constituents from C2-units by a modified tricarboxylic acid cycle. Nature 179: 988–991.
Leigh JA, Dodsworth JA . (2007). Nitrogen regulation in bacteria and archaea. Annu Rev Microbiol. 61: 349–377.
Liu H, Han J, Liu X, Zhou J, Xiang H . (2011a). Development of pyrF-based gene knockout systems for genome-wide manipulation of the archaea Haloferax mediterranei and Haloarcula hispanica . J Genet Genomics 38: 261–269.
Liu H, Wu Z, Li M, Zhang F, Zheng H, Han J et al. (2011b). Complete genome sequence of Haloarcula hispanica, a model haloarchaeon for studying genetics, metabolism, and virus-host interaction. J Bacteriol 193: 6086–6087.
Malki L, Yanku M, Borovok I, Cohen G, Mevarech M, Aharonowitz Y . (2009). Identification and characterization of gshA, a gene encoding the glutamate-cysteine ligase in the halophilic archaeon Haloferax volcanii . J Bacteriol 191: 5196–5204.
Nelson-Sathi S, Dagan T, Landan G, Janssen A, Steel M, McInerney JO et al. (2012). Acquisition of 1,000 eubacterial genes physiologically transformed a methanogen at the origin of Haloarchaea. Proc Natl Acad Sci USA 109: 20537–20542.
Nelson-Sathi S, Sousa FL, Roettger M, Lozada-Chávez N, Thiergart T, Janssen A et al. (2014). Origins of major archaeal clades correspond to gene acquisitions from bacteria. Nature 517: 77–80.
Oren A . (1995). Uptake and turnover of acetate in hypersaline environments. FEMS Microbiol Ecol 18: 75–84.
Oren A . (2002) Halophilic Microorganisms and Their Environments. Kluwer Academic Publishers: : Dordrecht, Netherland.
Oren A, Gurevich P . (1995). Isocitrate lyase activity in halophilic archaea. FEMS Microbiol Lett 130: 91–95.
Quillaguamán J, Guzmán H, Van-Thuoc D, Hatti-Kaul R . (2010). Synthesis and production of polyhydroxyalkanoates by halophiles: current potential and future prospects. Appl Microbiol Biotechnol 85: 1687–1696.
Reeves HC, O’Neil S, Weitzman PD . (1983). Modulation of isocitrate dehydrogenase activity in Acinetobacter calcoaceticus by acetate. FEBS Lett 163: 265–268.
Reeves HC, O’Neil S, Weitzman PD . (1986). Changes in NADP-isocitrate dehydrogenase isoenzymes levels in Acinetobacter calcoaceticus in response to acetate. FEMS Microbiol Lett 35: 229–232.
Riddles PW, Blakeley RL, Zerner B . (1983). Reassessment of Ellman’s reagent. Methods Enzymol 91: 49–60.
Sasikaran J, Ziemski M, Zadora PK, Fleig A, Berg IA . (2014). Bacterial itaconate degradation promotes pathogenicity. Nat Chem Biol 10: 371–377.
Serrano JA, Bonete MJ . (2001). Sequencing, phylogenetic and transcriptional analysis of the glyoxylate bypass operon (ace in the halophilic archaeon Haloferax volcanii . Biochim Biophys Acta 1520: 154–162.
Serrano JA, Camacho M, Bonete MJ . (1998). Operation of glyoxylate cycle in halophilic archaea: presence of malate synthase and isocitrate lyase in Haloferax volcanii . FEBS Lett 434: 13–16.
Simon EJ, Shemin D . (1953). The preparation of S-succinyl coenzyme-A. J Am Chem Soc 75: 2520.
Stadtman ER . (1957). Preparation and assay of acyl coenzyme A and other thiol esters; use of hydroxylamine. Methods Enzymol 3: 931–941.
Strijbis K, Distel B . (2010). Intracellular acetyl unit transport in fungal carbon metabolism. Eukaryot Cell 9: 1809–1815.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S . (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729.
Thompson JD, Higgins DG, Gibson TJ . (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680.
Williams D, Gogarten JP, Papke RT . (2012). Quantifying homologous replacement of loci between haloarchaeal species. Genome Biol Evol 4: 1223–1244.
Zarzycki J, Brecht V, Müller M, Fuchs G . (2009). Identifying the missing steps of the autotrophic 3-hydroxypropionate CO2 fixation cycle in Chloroflexus aurantiacus . Proc Natl Acad Sci USA 106: 21317–21322.
Acknowledgements
This work was funded by the Deutsche Forschungsgemeinschaft (BE 4822/3-1 and Heisenberg fellowship to IAB) and partially by the National Natural Science Foundation of China (Grant Nos. 31330001 to HX). We thank G Fuchs, Freiburg, for discussions, suggestions and critical reading of the manuscript, and N Gad’on and C Ebenau-Jehle, Freiburg, for maintaining the laboratory.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Borjian, F., Han, J., Hou, J. et al. The methylaspartate cycle in haloarchaea and its possible role in carbon metabolism. ISME J 10, 546–557 (2016). https://doi.org/10.1038/ismej.2015.132
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2015.132
This article is cited by
-
Synthetic anaplerotic modules for the direct synthesis of complex molecules from CO2
Nature Chemical Biology (2023)
-
Enoyl-CoA hydratase mediates polyhydroxyalkanoate mobilization in Haloferax mediterranei
Scientific Reports (2016)