Abstract
Highly diverse microbial assemblages colonize plant roots. It is still poorly understood whether different members of this root microbiome act synergistically by supplying different services (for example, different limiting nutrients) to plants and plant communities. In order to test this, we manipulated the presence of two widespread plant root symbionts, arbuscular mycorrhizal fungi and nitrogen-fixing rhizobia bacteria in model grassland communities established in axenic microcosms. Here, we demonstrate that both symbionts complement each other resulting in increased plant diversity, enhanced seedling recruitment and improved nutrient acquisition compared with a single symbiont situation. Legume seedlings obtained up to 15-fold higher productivity if they formed an association with both symbionts, opposed to productivity they reached with only one symbiont. Our results reveal the importance of functional diversity of symbionts and demonstrate that different members of the root microbiome can complement each other in acquiring different limiting nutrients and in driving important ecosystem functions.
Similar content being viewed by others
Introduction
Microbes have key roles in ecosystems and influence a large number of important ecosystem processes, including plant nutrient acquisition, nitrogen and carbon cycling, and soil formation (Wardle et al., 2004; Van der Heijden et al., 2008; Van Elsas et al., 2012; Wagg et al., 2014). While it is widely recognized that microbes perform crucial roles in biogeochemical cycling, the impact of underground root-associated microbes on plant productivity and plant biodiversity is less well understood. This is due to their hidden lifestyle, the fact that many microbes cannot be cultivated (Curtis et al., 2002), and because it is difficult to manipulate the presence and diversity of microbes without contamination (Read, 2002). We refer to root-associated microbes (bacteria and fungi) as symbionts following De Bary’s (1879) definition of symbiosis as the living together of two or more unlike organisms. We use the term symbiont for the microbial partner of the host plant without qualifying to the nature of the relationship (mutualistic, commensalistic or parasitic).
Earlier work showed that an abundant group of plant symbionts, the arbuscular mycorrhizal (AM) fungi, enhanced plant diversity and plant productivity (Grime et al., 1987; Van der Heijden et al., 1998). However, besides phosphorus acquiring AM fungi, plants associate with a wide range of other microbial symbionts, including nitrogen-fixing bacteria, endophytic fungi and bacteria, and a large range of microbes with unknown functions awaiting discovery (Van der Heijden et al., 2008; Drigo et al., 2010; Mendes et al., 2011; Bulgarelli et al., 2012; Sessitsch et al., 2012). These microbial symbionts form complex and highly diverse communities, also called the root or rhizosphere microbiome (Mendes et al., 2011; Berendsen et al., 2012; Bulgarelli et al., 2013). The interactive effects of different microbial symbionts on plants and plant communities are only partly understood as most studies tested the interactive effects of different microbial symbionts on single plants or plants grown in monoculture (for example, Requena et al., 1997; Mack and Rudgers, 2008; Larimer et al., 2010). We hypothesized that the interaction of different groups of symbionts enhances plant productivity and plant diversity since they can act synergistically (effects due to interaction become larger compared with the addition of single effects). Interacting symbionts may stimulate ecosystem functioning by supplying different services (for example, complementary limiting nutrients) to plants and by supporting different plant species. This also supports the biodiversity hypothesis that two organisms (or groups of organisms) enhance productivity and ecosystem functioning by utilizing different resource pools via their differentiation in life strategy (Tilman et al., 1997).
In this study, we focus on two abundant and widespread groups of microbial symbionts: phosphorus acquiring AM fungi and nitrogen-fixing rhizobia bacteria (hereafter Rhizobia). AM fungi associate with about 70% of all land plants, are dominant in a wide range of ecosystems, and facilitate plant nutrient uptake, especially phosphorus (Smith and Read, 2008; Bonfante and Genre, 2010; Van der Heijden et al., 2015). Rhizobia associate with legumes and fix atmospheric nitrogen into plant available ammonium. Associations between legumes and rhizobia are vital for maintaining plant productivity in a wide range of ecosystems (Cleveland et al., 1999). Hence, these observations suggest that both symbionts have key roles in natural ecosystems because they enhance the availability of phosphorus and nitrogen, the two most important elements that limit plant productivity (Elser et al., 2007).
Legumes are the most important contributors to biological nitrogen fixation in terrestrial ecosystems, which totals approximately 128 Tg of N per year (Cleveland et al., 1999; Galloway et al., 2004). Large amounts of nitrogen are fixed by legumes in widespread vegetation types such as tropical savanna (for example, by various Acacia species), arid shrub land (for example, by mesquite) and in tropical rain forest (for example, various Inga spp., Cleveland et al., 1999; De Faria et al., 2010). Nitrogen fixation by legumes is also high in agricultural systems (for example, soybean) totalling approximately 18.5 Tg of N per year (Herridge et al., 2008). About 95% of all legumes, including well-known and highly abundant species such as Acacia sp., Arachis hypogaea, Glycine max, Prosopis sp., Medicago sativa, Mimosa sp., Phaseolus sp., Pisum sativum and Trifolium sp., form a symbiosis with AM fungi (Wang and Qiu, 2006). Hence, AM fungi may indirectly contribute to the global N cycle by providing phosphorus and other nutrients to N-fixing legumes, especially in natural ecosystems that are often P limited (Elser et al., 2007).
Glasshouse and field experiments with agricultural legumes (for example, soybean, bean and clover) suggest that tripartite symbiotic associations among legumes, AM fungi and rhizobia do not necessarily enhance plant yield (for example, Mosse et al., 1976; Bethlenfalvay et al., 1982; Siviero et al., 2008; Larimer et al., 2010). However, host plant services of AM fungi and rhizobia are expected to be much higher under conditions where nutrient availability is low such as in most natural ecosystems. A recent study suggested that phosphatase enzymes and increased root colonization levels by AM fungi enhance the capacity of legumes to acquire soil P and prosper in tropical forest (Nasto et al., 2014). Experimental proof for the interactive effects of AM fungi and rhizobia on plant communities is missing due to experimental constraints (rhizobial contamination in control treatments and the availability of an experimental system to evaluate microbial interactions in plant communities under controlled conditions) and it is unclear whether AM fungi and rhizobia jointly increase legume growth. Moreover, so far, most biologists viewed legumes as important ecosystem engineers due to their ability to associate with nitrogen-fixing rhizobia. However, it has rarely been considered that besides rhizobia, AM fungi may be instrumental to the success of legumes in a wide range of ecosystems.
In this study, the contribution of an AM fungus and rhizobia bacteria to ecosystem functioning (here: perennial grassland) was examined by quantifying plant productivity, nutrient uptake, plant community structure, plant diversity and seedling establishment. For this purpose, we reconstructed the plant community (Table 1) of a Dutch dune grassland reference site (Supplementary Figure S1) in gnotobiotic experimental microcosms (Supplementary Figure S2). These microcosms contained the native soil, and native plants and microbial taxa collected from the reference site. We inoculated an AM fungus and rhizobia bacteria alone or in combination in the microcosms and control microcosms without AM fungus and rhizobia were also set up. We successfully manipulated the microbial symbionts in the microcosms without contamination from the outside until the end of the experiment after sixteen-and-a-half months (see Supplementary Information, Supplementary Table S1). We added labeled nitrogen to the microcosms enabling us to estimate the amount of biologically fixed nitrogen. To test effects of AM fungi and rhizobia on seedling recruitment in established vegetation, we planted seedlings of seven grassland species into 11-month-old microcosms between the existing vegetation. This enabled us to simulate seedling establishment in established plant communities. We hypothesized that:
(1) Both symbionts complement each other resulting in enhanced plant diversity, nutrient acquisition and plant productivity compared with a single symbiont situation.
(2) Legume biomass is higher in the presence of the bacterial and fungal symbionts compared with a single symbiont situation.
(3) The presence of symbionts facilitates seedling establishment of responsive plants and the establishment is more efficient in the presence of both symbionts compared with single symbiont situations.
Materials and methods
Experimental microcosms and biological material
We reconstructed a Dutch dune perennial grassland ecosystem by using the AM fungi, rhizobia and plant species that all co-occur at ‘Noordhollands Duinreservaat’ (Supplementary Figure S1; Egmond Binnen, 52° 40’N 4° 39’E), to which we refer to as the reference site. Thirty-six microcosms (Supplementary Figure S2) were established under sterile conditions in closed growth chambers (for further details: Supplementary Information,Wagg et al., 2014).
The experiment was set up as a full factorial design with four treatments of nine replicates each. The microcosms were inoculated with the following treatments: with AM fungi (M), with rhizobia (R), with AM fungi and rhizobia (MR) or controls without symbionts (C). Microcosms received 350 spores of the AM fungus Rhizoglomus irregularis (isolate BEG 21, Genbank accession number DQ377990; previously named Glomus intraradices and Rhizophagus irregularis, Sieverding et al., 2014). This AM fungus has a worldwide distribution, and is common in Dutch dune grasslands including the reference site (Scheublin et al., 2004; Scheublin and van der Heijden, 2006). The spores of this fungus were placed approximately 5 cm below the soil surface when assembling the microcosms. The spores were surface sterilized by an ultrasound treatment (4 times 10 s at 40 kHz), followed by a 12-h treatment with a filter-sterilized antibiotic solution (Streptomycin 200 mg l−1 and Gentamycin 100 mg l−1) and a 10-min treatment with a 2% Chloramine-T solution mixed with a few drops of Tween-20 (procedure modified after Bécard and Piché, 1992), while non-mycorrhizal microcosms received autoclaved (110 °C for 2 h) spores. Inoculation with rhizobia was conducted by means of a 25-ml water suspension, which contained equal amounts of four rhizobia strain suspensions with an OD580 of 0.1 (R microcosms), while the microcosms without rhizobia received an autoclaved (121 °C for 41 min) suspension. The rhizobia strains were isolated from root nodules of Lotus corniculatus L. (two isolates: L.c. 2 and L.c. 3) and Trifolium repens L. (two isolates: T.r. 1 and T.r. 2) as described previously (see Van der Heijden et al. 2006 for details).
Seedlings of 11 plant species (Table 1) were planted in each microcosm, representative for three common plant functional groups at the reference site. Seeds were collected from or near the reference site, except for T. repens seeds, which were obtained from a commercial company that supplies seeds originating from natural populations in The Netherlands (Cruydt-hoeck, Nijeberkoop, The Netherlands). Seeds were surface sterilized (using a 5% commercial bleach solution, with varying adaptations depending on plant species) and allowed to germinate on 1.6% water agar (legumes) or in autoclaved sand (non-legume plant species). Each microcosm had a unique planting design, where the location of the seedlings was determined through randomization. Five weeks after the initial establishment of the plant communities, seedlings that did not survive or appeared unhealthy were replaced by seedlings of the same age.
Measurements and data analysis
Five-and-a-half and eleven months following the initial start of the experiment, aboveground living and dead plant material was clipped at approximately 2 cm above the soil surface. This treatment was performed to simulate hay making or grazing, which is commonly employed at natural dune grasslands comparable to the reference site. Two of the investigated plant species, Hieracium and Carlina, grow close to the soil surface and therefore produced only little or no biomass above 2 cm at the first or second harvest. Final harvest of the microcosms occurred after sixteen-and-a-half months.
At each harvest, the shoots were harvested split by plant species, oven dried and weighed (Supplementary data 1). Unless otherwise stated, the biomass data of the three harvests were combined per plant species to determine each plant’s productivity in the grassland microcosms. Plant productivities per functional group or total plant productivity of a microcosm (shoot and root data) were calculated by summing up the respective individual dry weights. At final harvest, the plant survival was determined for each plant and functional group (Supplementary data 2). Shannon’s diversity index H was calculated for each microcosm using the shoot biomass per plant species in the microcosm (sum of the three harvests) as the surrogate for species abundance in the equation (see Supplementary Data 5 for the R code).
Roots (all plant species together) were collected from the soil, divided into subsamples and used to determine root biomass, root length and the percentage of root length colonized by AM fungi (McGonigle et al., 1990). A soil subsample was used to determine hyphal length (Jakobsen et al., 1992). Small amounts of representative root material for each legume species were collected from the soil and checked for nodulation. Dried plant material of roots (all plant species together) and of shoots of each plant species individually was ground in a ball mill and used to determine phosphorus concentrations (Murphy and Riley, 1962). Nitrogen concentrations and the concentration of the nitrogen isotope 15N (measured as δ15N) of plant material were determined using an elemental analyzer (NC2500; ThermoQuest Italy, Rodana, Italy), coupled with a continuous-flow isotope mass spectrometer (DeltaPlus; ThermoQuest Finnigan, Bremen, Germany). Subsequently, total N content and P content were determined for total plant communities and shoot N content and P content were determined per plant functional group (grasses, herbs and legumes; Supplementary data 3). The percentage of symbiotically fixed N (Nfix) of the plant communities was estimated after (Högberg, 1997) using δ15N values of the plant communities:

where δ15Nsym is the average δ15N value of plant N in plant communities with one or both symbionts and δ15Nnonsym is the average δ15N of plant N in microcosms without a symbiont, which was 1036‰. A is the δ15N value of a plant community completely dependent on atmospheric nitrogen and was assumed to be zero in this study. The δ15N value of each plant community was calculated based on a mass balance of the observed δ15N values in the concerning plant species and roots.
Seedling establishment
Eleven months after the initial start of the experiment, 5–7 days old seedling were planted into the microcosms at specified positions immediately after the cutting of the mature plant community following earlier work (Grime et al., 1987). The planting of the seedlings simulates a ‘controlled’ seed rain, which is natural in many grasslands. Seedlings of two plant species per functional group were included (Table 1): the legume species L. corniculatus, and T. repens, the grasses Festuca ovina and Koeleria macrantha, and the herbs Plantago lanceolata and Senecio jacobaea. These plant species were also planted at the start of the experiment and are all perennial plant species. In addition, seedlings of the two annual legume species Trifolium dubium Sibth. and Trifolium arvense L. were planted. These annual plant species, which are also common at the reference site, were not added at the start of the experiment, as we wanted to establish a perennial plant community first. Moreover, we choose two additional legumes because we wanted to test, following observation on L. corniculatus and T. repens, whether other legume species also depend on both symbionts for optimal performance. Both Rhizobium strains isolated from T. repens nodules were able to induce root nodules on T. arvense and T. dubium when grown in isolation with these plant species (data not shown). At the final harvest of the microcosms, five-and-a-half months after planting the seedlings, the dry shoot biomass of each seedling was determined (Supplementary data 4).
Experimental design and statistical analysis
The experiment was set up as a complete randomized block design with two factors AM fungi (with or without AM fungi) and rhizobia (with or without rhizobia). The four treatments were replicated nine times where each replicate was assigned to one block, making a total of nine blocks.
Repeated biomass measurements were analyzed for all 11 plant species and for the total shoot biomass using the linear effect models using the R-package ‘nlme’ (Pinheiro et al., 2009). The linear mixed effect models for shoot biomass included measurement time (biomass data of harvest 1, 2 and 3), AMF treatment, Rhizobia treatment and their interactions as fixed effects. Significant block effects did not occur for any of the variables (Supplementary Table S3) and, therefore, no correction for block effects was made in the subsequent two-way analyses of variance (ANOVA).
ANOVA was conducted for all measured variables using the function ‘aov‘ in modeling the factors Rhizobia, Mycorrhiza and their interaction (Supplementary Table S1). We subsequently conducted pair-wise comparisons between the treatments for the Rhizobia × Mycorrhiza interaction using Tukey’s HSD (honestly significant difference) post hoc test. A one-way ANOVA was conducted on the percentage of root colonization by AM fungi to compare the two treatments with AM fungi because the non-mycorrhizal treatments remained free of mycorrhizal structures. Variables were examined for normality and homogeneity of variances using the Shapiro-Wilk and Bartlett tests, respectively, and if necessary, natural log or rank transformed to meet the requirements to perform the ANOVA.
To assess the influence of symbionts on the observed variation in biomass production of the individual plant species, we performed a constrained correspondence analysis, a.k.a. canonical correspondence analysis. Constrained correspondence analysis differs from unconstrained correspondence analysis that the ordination is constrained such that the variation attributable to the factors AM fungi and rhizobia is displayed. We employed the function ‘cca‘ of R package ‘vegan‘ (Oksanen et al., 2013).
Statistical analyses were performed in R (R Core Team R, 2014; version 3.1.1, 2014-07-10) and custom R scripts related to the statistical analysis and plotting of the figures are made available as Supplementary data 5.
Results
Plant productivity, diversity and community composition
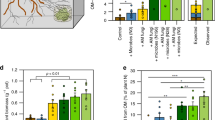
Plant community composition of the grassland communities depended strongly on the presence or absence of both symbionts (Figure 1a) and the different plant functional groups (grasses, herbs and legumes) responded differentially to the treatments. The grasses produced the majority of the biomass in the microcosms when AM fungi were absent. In contrast, herbs and legumes dominated the microcosms where AM fungi were present. The total productivity of the microcosms (shoot and roots combined) varied only little between the treatments (Figure 1a) while the interaction of AM fungi and rhizobia affected the plant productivity positively (Supplementary Table S1). Plant diversity was positively affected by AM fungi and not by rhizobia (Supplementary Table S1). The presence of the AM fungi in combination with rhizobia enhanced plant diversity in the microcosms (Figure 1b). The presence of AM fungi alone tended to augment plant diversity while rhizobia alone did not affect plant diversity. In summary, the presence and absence of bacterial and fungal symbionts affected the plant species composition but not the productivity of the microcosms. Hence, different plant species accounted for the produced biomass in the microcosms.
Productivity and diversity of microcosms. The productivity of the microcosms (a) is reported with the dry weight shoot biomass (sum of the three harvests) for the three plant functional groups (legumes, herbs and grasses) and the biomass of the roots at the third harvest. Plant diversity (b) is based on the shoot biomass data of the three harvests. Grassland microcosms contained no plant symbionts (C), only rhizobia (R), only AM fungi (M) or both symbionts (MR). Bars represent means (n=9; ±s.e.m.) and letters (in a are colored by functional group) indicate statistical significance; treatments not sharing a letter differ at P<0.05 (Tukey’s HSD). The total productivity of the microcosms (shoot and roots combined) was not significantly different among the treatments. Statistic details are given in Supplementary Table S1.
The productivity of the individual plant species depended strongly on whether AM fungi and or rhizobia were present or absent (Figure 2). Clustering analysis revealed two groups of plant species, those benefitting from the presence of AM fungi (the herbs and the legumes) and those obtaining highest biomass in microcosms without AM fungi (the three grasses and the wood rush Luzula campestris). Moreover, within the group of plant species responding positively to AMF, we observed that the biomass of the two legumes, L. corniculatus and T. repens, was highest in the combined treatment of AM fungi and rhizobia compared with the single symbiont treatments (Supplementary Table S2).
Plant community composition. The heatmap reports the dry weight shoot biomass of individual plant species (in rows) in all microcosms (in columns). The data comprise the summed productivity of the three harvests and are presented with a log scale. Dendrograms report similarities in productivity of the different plant species (left of the heatmap) and in the differentially treated microcosms (above the heatmap) based on complete linkage clustering using Pearson distances. Microcosms containing no plant symbionts (C), only rhizobia (R), only AM fungi (M), or both symbionts (MR) as well as the plant functional groups are marked as indicated.
The plants in the microcosms were harvested three times (we did this to simulate hay making or grazing, which is a common practice in dune grassland). Subsequently, we examined the variation in biomass production of each plant species over time using linear mixed model analysis (Supplementary Table S3). We found a significant harvest effect for each plant species (Supplementary Table S3) as evidenced with the increasing productivity in the grassland microcosms over time, being lowest at the first harvest and highest at the third harvest (Supplementary Figure S3).
Plant survival
The survival rate of the various plant species was strongly affected by the presence or absence of the symbionts (Supplementary Figure S4; Supplementary Table S1) with differential responses for each functional group of plant species. Grasses had higher survival rates (82.9%) compared with herbs and legumes (7.2% and 12.4%, respectively) when AM fungi were absent. But when AM fungi were present the herbs and legumes had much higher survival rates (above 75%). The presence of rhizobia in the microcosms positively affected the survival rate of the legumes (Supplementary Table S1).
Microbial influence on plant community composition
To approximate the contribution of AM fungi and the rhizobia bacteria to the observed variation in biomass production of the individual plant species, we employed constrained correspondence analysis. The analysis revealed that 75.5% (P<0.005; 95% confidence interval=22%, 182%) of the variation in biomass production across the microcosms could be attributed to the presence of the symbionts (Supplementary Figure S5). The majority (95.6%) of this variation was explained by the presence or absence of AM fungi resulting in a clear separation of non-mycorrhizal relative to mycorrhizal microcosms along the first axis. The presence or absence of rhizobia explained 3.9% of the variation, and resulted in separate clusters within the mycorrhizal microcosms along the second axis. Fitting the plant species as vectors onto the ordination plot confirmed the functional group-wise responses of the plants to the AMF and rhizobia treatments: grasses pointed to the non-mycorrhizal microcosms while the herbs and legumes are directed toward mycorrhizal microcosms, where the latter were bent to the microcosms also containing rhizobia. Overall, we concluded that mainly the AM fungi and to minor extent rhizobia explained the differences in biomass production of the individual plant species and that these plant growth responses followed the logic of the plant functional groups.
Plant nutrient acquisition
We determined plant phosphorus and plant nitrogen acquisition in each microcosm based on the total plant biomass (shoot and roots). AM fungi enhanced plant phosphorus uptake (Figure 3a; Supplementary Table S1), while rhizobia in the presence of AM fungi enhanced plant nitrogen uptake (Figure 3b; Supplementary Table S1). The estimated fraction of biologically fixed nitrogen was on average 26% (77.8±6.9 mg fixed N) in microcosms inoculated with AM fungi and with rhizobia and only 3% (6.1±0.74 mg fixed N) in microcosms with rhizobia but without AM fungi. This demonstrated that both symbionts were needed for maximum nitrogen fixation. The plant nitrogen content that was not derived from biological nitrogen fixation (the mineral N content) was similar in all four treatments suggesting that the presence or absence of AM fungi did not affect the mineral plant N uptake (Figure 3b).
Plant nutrient acquisition. P content (a) and mineral and symbiotically fixed N content (b) in plants of the grassland microcosms containing no plant symbionts (C), only rhizobia (R), only AM fungi (M), both symbionts (MR). Total plant nutrient content (shoot and roots combined) refers to the overall plant nutrient uptake in the microcosms (sum of the three harvests). Bars represent means (n=9;±s.e.m.) and letters indicate statistical significance; treatments not sharing a letter differ at P<0.05 (Tukey’s HSD). Statistic details and the nutrient acquisition data per functional groups are reported in Supplementary Table S1 and Supplementary Figure S6, respectively.
The distribution of above ground phosphorus and shoot nitrogen among the coexisting plant functional groups varied greatly depending on whether AM fungi, rhizobia or their combination was present (Supplementary Figure S6, Supplementary Table S1). In the absence of AM fungi, the majority of above ground phosphorus was present in grasses. However, when AM fungi were present, most above ground phosphorus was found in herbs and legume shoot phosphorus content increased up to 65.6-fold (Supplementary Table S1). Moreover, the fraction of nitrogen found in legume shoots was low (on average 3.1%) in treatments without symbionts or with only one symbiont. However, when both symbionts were present 31.5% of above ground nitrogen was found in legumes.
Seedling establishment
The grassland microcosms were further utilized to test whether seedling establishment of a new generation of plants depends on the presence of one or both symbionts. We specifically asked whether the establishment response differs among plant species by their functional group. We employed two plant species per functional group (Table 1), all perennial plant species that were already planted at the start of the experiment, and in addition the two annual legumes T. dubium and T. arvense were added. These two annual legume species also occur natively at the reference site. Seedlings were planted between the established plant communities eleven months after setting up the microcosms and immediately after cutting the mature grassland community. This was done to mimic natural seedling establishment. Seedling establishment was scored after 5.5 months at final harvest by quantifying shoot biomass. Seedlings of the four investigated legume species (L. corniculatus, T. arvense, T. dubium and T. repens) performed poorly in control microcosms or in microcosms with only AM fungi or only rhizobia. However, when both AM fungi and rhizobia were present the average biomass of L. corniculatus, T. arvense, T. dubium and T. repens seedlings increased respectively by 5.8-, 15.7-, 21.3- and 16.8-fold, compared with microcosms with only one symbiont (Figure 4,Supplementary Table S1). This demonstrates that both symbionts are required for the two annual and the two perennial legumes to establish successfully. In contrast to the legumes, the seedlings of the grasses (F. ovina and K. macrantha) and the herbs (P. lanceolata and S. jacobaea) did not or only marginally respond to the presence of AM fungi and/or rhizobia.
Seedling establishment. After 11 months, seedlings were added between the plants, i.e., into the established grasslands microcosms containing no plant symbiont (C), only rhizobia (R), only AM fungi (M), or both symbionts (MR). Seedling establishment was approximated for each developing plant individual with the shoot dry weight of the biomass harvested 5.5 months later at the third harvest. Plant species and their functional groups are indicated. Bars represent means (n=9; ±s.e.m.), letters below the bars indicate statistical significance; treatments not sharing a letter differ at P<0.05 (Tukey’s HSD) and statistical details are given in Supplementary Table S1.
Discussion
This study demonstrates that a widespread plant-fungal-bacterial symbiosis promotes plant diversity and seedling recruitment of legumes in microcosms simulating European grassland. Plant diversity was only higher in grassland microcosms where both AM fungi and rhizobia were present compared with microcosms without symbionts, pointing to the importance of both symbionts as a driver of diversity in grassland. Interestingly, seedlings of the four studied legume species all relied on the presence of both AM fungi and rhizobia to be successful in the grasslands as shown by a 5.8- up to 14.8-fold higher biomass in microcosms where both symbionts are present compared with microcosms inoculated with only rhizobia. Similarly, the total biomass of legumes was enhanced in microcosms where both symbionts were present compared with a single symbiont situation. It is well known that rhizobia have a key role in natural and agricultural ecosystems by fixing nitrogen and promoting plant productivity (Galloway et al., 2004; Elser et al., 2007). Earlier work also showed that AM fungi contribute significantly to plant diversity in European calcareous grasslands (Grime et al., 1987; Van der Heijden et al., 1998). Moreover, several studies, mainly performed with agricultural legumes, tested the interactive effects of AM fungi and rhizobiaon plant growth (Requena et al., 1997; Larimer et al., 2010). However, the interactive effects of both symbionts and their influence on plant community composition, N fixation and P uptake in nutrient poor grassland communities had not been conclusively shown before.
Especially interesting is the strong reliance of legumes on the combined presence of both rhizobia bacteria and AM fungus. The use of our growth chambers, designed to prevent microbial contamination from the outside, made it possible to test the interactive effects of both microbial symbionts and to successfully manipulate the presence of AM fungus and rhizobia for more than one growing season without contamination. Subsequently, we showed that the legumes performed much better when both symbionts were present, while legume growth, especially of the seedlings, was lower or negligible when only one symbiont (either rhizobia bacteria or AM fungus) was present. Our results are likely to be especially relevant for natural ecosystems such as species rich grassland where soil phosphorus availability is often low (for example, Janssens et al., 1998; Elser et al., 2007). Most legumes have a high phosphorus requirement (Scheublin and van der Heijden, 2006). Thus, the observed enhanced phosphorus uptake by extensive mycorrhizal hyphal networks and subsequent transfer of phosphorus to legumes likely facilitated legume growth and biological nitrogen fixation in this nutrient poor grassland community. Our finding complements a recent study (Nasto et al., 2014), which showed that nitrogen fixing tropical legumes have higher AM fungal root colonization levels and enhanced root phosphatase activity compared with non-fixers. The positive effect of AM fungi on nitrogen fixation in our study is probably indirect, in that AM fungi supply phosphorus to phosphorus limited legume roots that subsequently can facilitate and enhance nitrogen fixation by rhizobia in the root nodules. Direct effects on biological nitrogen fixation are unlikely because in earlier work we showed that root nodules, colonized by AM fungi, did not fix nitrogen (Scheublin and van der Heijden, 2006).
It is difficult to assess the impact of AM fungi and rhizobia on plant diversity and ecosystem processes in the field because AM fungi and rhizobia are usually already present in the soil and their abundance cannot be easily manipulated without simultaneously changing other factors or organisms (Read, 2002). The attempts that have been made to test interactive effects of various groups of soil microorganisms on plant communities often suffer from contamination in various treatments or focus on single plant species (Requena et al., 1997; Mack and Rudgers, 2008; Bauer et al., 2012). In this study, microcosms were established under controlled conditions in sterilized soil and in specially designed mini growth chambers where incoming air and water was filtered to prevent microbial contamination. In this way, it was possible, for the first time, to successfully manipulate the presence of AM fungi and rhizobia in plant communities without contamination for over 1 year (16.5 months). It is, however, important to consider that this study design in model systems with sterilized soil, the absence of larger soil organisms and manipulation in microcosm under greenhouse conditions has limitations (see Read, 2002 for a discussion). To avoid these limitations as much as possible the soil and organisms (rhizobia, plant species) used for this experiment were all collected from the same dune grassland area and all organisms naturally coexist. In order to be able to make general conclusions and perform meta-analyses, future studies should test the importance of symbiont diversity for plants and plant communities using different soils, plant communities and legume species. Such studies should also include a control treatment where a full soil community (with non-sterilized soil as inoculum) is added to the microcosms. Moreover, in this study, we used only one AMF taxa (Rhizoglomus irregularis), and future studies should include a higher number of AMF taxa (both from the same and from different AMF species) to enhance genetic and functional diversity, which is prevalent in the field (Vandenkoornhuyse et al., 2003; Scheublin et al., 2004; Munkvold et al., 2004).
The composition of the plant communities in the microcosms resembles those found in nutrient poor European grassland and the above ground productivity after 1 year (125–205 g m−2) is in the range found in nutrient poor, species rich natural grassland in the Netherlands (for example, Willems et al., 1993; Olde Venterink et al., 2003) and Western Europe (for example, Al-Mufti et al., 1977). Plant productivity in such grassland is low and strongly nutrient limited (Willems et al., 1993; Kooijman et al., 1998) compared with other grasslands, including fertilized ones where it is not unusual to have productivity of over 1000 g m−2 year-1) (e.g., Olde Venterink et al., 2003). Thus, under such nutrient poor conditions, plant symbionts are expected to be most important. Hence, the big effects of AM fungi and rhizobia on plant community composition and on the success of individual plants species (for example, the legumes) reported in this study probably reflects an upper limit and are to be expected in nutrient poor ecosystems, not in fertilized or nutrient-rich ecosystems.
In recent years, the role of plant diversity and individual plant species for regulating ecosystem functioning and stability has been stressed (Isbell et al., 2011; Hautier et al., 2014). Ecologists are becoming increasingly aware of the role of aboveground–belowground relationships in controlling ecosystem processes and properties (Wardle et al., 2004; Scherber et al., 2010; Eisenhauer, 2012; Wagg et al., 2014). A range of underground factors that determine aboveground plant community structure have now been identified, including soil disturbance, soil pathogens, mycorrhizal fungi, root herbivores and temporal and spatial variation in resource availability and activities of soil biota (for example, Wardle et al., 2004; Van der Heijden et al., 2008; Wagg et al., 2014). Our results show that, in addition, a widespread belowground plant-fungal-bacterial symbiosis has to be considered as an important determinant of plant productivity and plant community structure above ground.
Overall, our results emphasize the importance of diverse belowground symbionts as mediators of plant biodiversity and nutrient supply in nutrient poor soils. Moreover, our results indicate that a tripartite interaction between AM fungi, rhizobia and legumes, and not the legume–rhizobia association as is often assumed, is a key for understanding legume success in natural ecosystems with poor nutrient availability. Finally, these results reveal that there is synergism in soil and that different microbial groups can complement each other, and facilitate plant diversity and plant growth by supplying different limiting nutrients.
Accession codes
References
Al-Mufti MM, Sydes CL, Furness SB, Grime JP, Band SR . (1977). A quantitative analysis of shoot phenology and dominance in herbaceous vegetation. J Ecol 65: 759–791.
Bauer JT, Kleczewski NM, Bever JD, Clay K, Reynolds HL . (2012). Nitrogen-fixing bacteria, arbuscular mycorrhizal fungi, and the productivity and structure of prairie grassland communities. Oecologia 170: 1089–1098.
Bécard G, Piché Y . (1992). Establishment of vesicular-arbuscular mycorrhiza in root organ culture: review and proposed methodology. In Norris JR, Read DJ, Varma AK eds Methods in Microbiology Vol. 24 Academic Press: London, pp 89–108.
Berendsen RL, Pieterse CM, Bakker PA . (2012). The rhizosphere microbiome and plant health. Trends Plant Sci 17: 478–486.
Bethlenfalvay GJ, Pacovsky RS, Bayne HG, Stafford AE . (1982). Interactions between nitrogen fixation, mycorrhizal colonization, and host-plant growth in the phaseolus-rhizobium-glomus symbiosis. Plant Physiol 70: 446–450.
Bonfante P, Genre A . (2010). Mechanisms underlying beneficial plant-fungus interactions in mycorrhizal symbiosis. Nat Commun 1: 48.
Bulgarelli D, Rott M, Schlaeppi K, Ver Loren van Themaat E, Ahmadinejad N, Assenza F et al. (2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488: 91–95.
Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P . (2013). Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64: 807–838.
Cleveland CC, Townsend AR, Schimel DS, Fisher H, Howarth RW, Hedin LO et al. (1999). Global patterns of terrestrial biological nitrogen (N2) fixation in natural ecosystems. Global Biogeochem Cycles 13: 623–645.
Curtis TP, Sloan WT, Scannell JW . (2002). Estimating prokaryotic diversity and its limits. Proc Natl Acad Sci USA 99: 10494–10499.
De Bary A . (1879) Die Erscheinung der Symbiose. Strassburg, Germany: Verlag von Karl J.Trubner.
De Faria SM, Diedhiou AG, de Lima HC, Ribeiro RD, Galiana A, Castilho AF et al. (2010). Evaluating the nodulation status of leguminous species from the Amazonian forest of Brazil. J Exp Bot 61: 3119–3127.
Drigo B, Pijl AS, Duyts H, Kielak AM, Gamper HA, Houtekamer MJ et al. (2010). Shifting carbon flow from roots into associated microbial communities in response to elevated atmospheric CO2. Proc Natl Acad Sci USA 107: 10938–10942.
Eisenhauer N . (2012). Aboveground-belowground interactions as a source of complementarity effects in biodiversity experiments. Plant Soil 351: 1–22.
Elser JJ, Bracken MES, Cleland EE, Gruner DS, Harpole WS, Hillebrand H et al. (2007). Global analysis of nitrogen and phosphorus limitation of primary producers in freshwater, marine and terrestrial ecosystems. Ecol Lett 10: 1135–1142.
Galloway JN, Dentener FJ, Capone DG, Boyer EW, Howarth RW, Seitzinger SP et al. (2004). Nitrogen cycles: Past, present, and future. Biogeochemistry 70: 153–226.
Grime JP, Mackey JML, Hillier SH, Read DJ . (1987). Floristic diversity in a model system using experimental microcosms. Nature 328: 420–422.
Hautier Y, Seabloom EW, Borer ET, Adler PB, Harpole WS, Hillebrand H et al. (2014). Eutrophication weakens stabilizing effects of diversity in natural grasslands. Nature 508: 521–525.
Herridge DF, Peoples MB, Boddey RM . (2008). Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil 311: 1–18.
Högberg P . (1997). Tansley Review NO 95 – N-15 Natural abundance in soil — plant systems. New Phytol 137: 179–203.
Isbell F, Calcagno V, Hector A, Connolly J, Harpole WS, Reich PB et al. (2011). High plant diversity is needed to maintain ecosystem services. Nature 477: 199–202.
Jakobsen I, Abbott LK, Robson AD . (1992). External hyphae of vesicular-arbuscular mycorrhizal fungi associated with Trifolium subterraneum L. New Phytol 120: 371–380.
Janssens F, Peeters A, Tallowin JRB, Bakker JP, Bekker RM, Fillat F et al. (1998). Relationship between soil chemical factors and grassland diversity. Plant Soil 202: 69–78.
Kooijman AM, Dopheide JCR, Sevink J, Takken I, Verstraten JM . (1998). Nutrient limitations and their implications on the effects of atmospheric deposition in coastal dunes; lime-poor and lime-rich sites in the Netherlands. J Ecol 86: 511–526.
Larimer AL, Bever JD, Clay K . (2010). The interactive effects of plant microbial symbionts: A review and meta-analysis. Symbiosis 51: 139–148.
Mack KML, Rudgers JA . (2008). Balancing multiple mutualists: Asymmetric interactions among plants, arbuscular mycorrhizal fungi, and fungal endophytes. Oikos 117: 310–320.
McGonigle TP, Miller MH, Evans DG, Fairchild GL, Swan JA . (1990). A new method which gives an objective measure of colonization of roots by vesicular–arbuscular mycorrhizal fungi. New Phytol 115: 495–501.
Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, JHM Schneider et al. (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332: 1097–1100.
Mosse B, Powell C, Hayman D . (1976). Plant growth responses to vesicular-arbuscular mycorrhiza. New Phytol 76: 331–342.
Munkvold L, Kjoller R, Vestberg M, Rosendahl S, Jakobsen I . (2004). High functional diversity within species of arbuscular mycorrhizal fungi. New Phytol 164: 357–364.
Murphy J, Riley JP . (1962). A modified single solution method for the determination of phosphate in natural waters. Anal Chim Acta 27: 31–36.
Nasto MK, Alvarez-Clare S, Lekberg Y, Sullivan BW, Townsend AR, Cleveland CC . (2014). Interactions among nitrogen fixation and soil phosphorus acquisition strategies in lowland tropical rain forests. Ecol Lett 17: 1282–1289.
Oksanen J, Blanchet F, Kindt R, Legendre P, Minchin P, O’Hara R et al. (2013). vegan: Community Ecology Package. R package version 2.0-10. R Packag version 1: 10.4135/9781412971874.n145.
Olde Venterink H, Wassen MJ, Verkroost AWM, De Ruiter PC . (2003). Species richness-productivity patterns differ between N-, P-, and K-limited wetlands. Ecology 84: 2191–2199.
Pinheiro J, Bates D, DebRoy S, Sarkar D R Core Team. (2009). nlme: Linear and Nonlinear Mixed Effects Models. R package version 3 1–120 Available at http://CRAN.R-project.org/package=nlme.
R Core Team. (2014) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria, http://www.R-project.org/.
Read DJ . (2002) Towards ecological relevance — progress and pitfalls in the path towards an understanding of mycorrhizal functions in nature. In: Van der Heijden MGA, Sanders IR (eds). Ecological Studies: Mycorrhizal Ecology, Vol. 157. Springer: Berlin, Heidelberg, pp 3–29.
Requena N, Jimenez I, Toro M, Barea JM . (1997). Interactions between plant-growth-promoting rhizobacteria (PGPR), arbuscular mycorrhizal fungi and Rhizobium spp. in the rhizosphere of Anthyllis cytisoides, a model legume for revegetation in mediterranean semi-arid ecosystems. New Phytol 136: 667–677.
Scherber C, Eisenhauer N, Weisser WW, Schmid B, Voigt W, Fischer M et al. (2010). Bottom-up effects of plant diversity on multitrophic interactions in a biodiversity experiment. Nature 468: 553–556.
Scheublin TR, Ridgway KP, Young JPW, Van Der Heijden MGA . (2004). Nonlegumes, legumes, and root nodules harbor different arbuscular mycorrhizal fungal communities. Appl Environ Microbiol 70: 6240–6246.
Scheublin TR, van der Heijden MGA . (2006). Arbuscular mycorrhizal fungi colonize nonfixing root nodules of several legume species. New Phytol 172: 732–738.
Sessitsch A, Hardoim P, Döring J, Weilharter A, Krause A, Woyke T et al. (2012). Functional characteristics of an endophyte community colonizing rice roots as revealed by metagenomic analysis. Mol Plant Microbe Interact 25: 28–36.
Sieverding E, da Silva GA, Berndt R, Oehl F . (2014). Rhizoglomus, a new genus of the Glomeraceae. Mycotaxon 129: 373–386.
Siviero MA, Motta AM, Lima D, dos S, Birolli RR, Huh SY, Santinoni IA et al. (2008). Interaction among N-fixing bacteria and AM fungi in Amazonian legume tree (Schizolobium amazonicum) in field conditions. Appl Soil Ecol 39: 144–152.
Smith SE, Read D . (2008) Mycorrhizal Symbiosis3rd edn. Academic Press: London, UK.
Tilman D, Knops J, Wedin D, Reich P, Ritchie M, Siemann E . (1997). The influence of functional diversity and composition on ecosystem processes. Science 277: 1300–1302.
Van der Heijden MGA, Bakker R, Verwaal J, Scheublin TR, Rutten M, van Logtestijn R et al. (2006). Symbiotic bacteria as a determinant of plant community structure and plant productivity in dune grassland. FEMS Microbiol Ecol 56: 178–187.
Van der Heijden MGA, Bardgett RD, Van Straalen NM . (2008). The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett 11: 296–310.
Van der Heijden MGA, Klironomos JN, Ursic M, Moutoglis P, Streitwolf-Engel R, Boller T et al. (1998). Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396: 69–72.
Van der Heijden MGA, Martin FM, Selosse M-A, Sanders IR . (2015). Mycorrhizal ecology and evolution: the past, the present, and the future. New Phytol 205: 1406–1423.
Van Elsas JD, Chiurazzi M, Mallon CA, Elhottova D, Kristufek V, Salles JF . (2012). Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc Natl Acad Sci 109: 1159–1164.
Vandenkoornhuyse P, Ridgway KP, Watson IJ, Fitter AH, Young JPW . (2003). Co-existing grass species have distinctive arbuscular mycorrhizal communities. Mol Ecol 12: 3085–3095.
Wagg C, Bender SF, Widmer F, van der Heijden MGA . (2014). Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proc Natl Acad Sci USA 111: 5266–5270.
Wang B, Qiu YL . (2006). Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza 16: 299–363.
Wardle DA, Bardgett RD, Klironomos JN, Setälä H, van der Putten WH, Wall DH . (2004). Ecological linkages between aboveground and belowground biota. Science 304: 1629–1633.
Willems JH, Peet RK, Bik L . (1993). Changes in chalk-grassland structure and species richness resulting from selective nutrient additions. J Veg Sci 4: 203–212.
Acknowledgements
We thank Richard Bardgett, Franz Bender, Wim van der Putten, Martin Stenflo, Herman Verhoef and Cameron Wagg for helpful comments that improved this manuscript. We are also grateful to Flip de Kriek and Fred Wolff for their technical support. This work was supported by the Netherlands Organisation for Scientific Research (NWO-Vernieuwingsimpuls; grant number 016.001.023), the Swiss National Science Foundation (SNF- Prodoc grant number 137136) and the Swiss Federal Government.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
van der Heijden, M., Bruin, S., Luckerhoff, L. et al. A widespread plant-fungal-bacterial symbiosis promotes plant biodiversity, plant nutrition and seedling recruitment. ISME J 10, 389–399 (2016). https://doi.org/10.1038/ismej.2015.120
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2015.120
This article is cited by
-
Effects of inoculating different mycorrhizal fungi on rhizosphere soil fungi and nutrient uptake of blueberry
Horticulture, Environment, and Biotechnology (2024)
-
Plant domestication shapes rhizosphere microbiome assembly and metabolic functions
Microbiome (2023)
-
Tree and shrub richness modifies subtropical tree productivity by regulating the diversity and community composition of soil bacteria and archaea
Microbiome (2023)
-
Identifying microbiota community patterns important for plant protection using synthetic communities and machine learning
Nature Communications (2023)
-
Dilution of specialist pathogens drives productivity benefits from diversity in plant mixtures
Nature Communications (2023)