Abstract
Permafrost-affected soils of the Siberian Arctic were investigated with regard to identification of nitrite oxidizing bacteria active at low temperature. Analysis of the fatty acid profiles of enrichment cultures grown at 4°C, 10°C and 17°C revealed a pattern that was different from that of known nitrite oxidizers but was similar to fatty acid profiles of Betaproteobacteria. Electron microscopy of two enrichment cultures grown at 10°C showed prevalent cells with a conspicuous ultrastructure. Sequence analysis of the 16S rRNA genes allocated the organisms to a so far uncultivated cluster of the Betaproteobacteria, with Gallionella ferruginea as next related taxonomically described organism. The results demonstrate that a novel genus of chemolithoautotrophic nitrite oxidizing bacteria is present in polygonal tundra soils and can be enriched at low temperatures up to 17°C. Cloned sequences with high sequence similarities were previously reported from mesophilic habitats like activated sludge and therefore an involvement of this taxon in nitrite oxidation in nonarctic habitats is suggested. The presented culture will provide an opportunity to correlate nitrification with nonidentified environmental clones in moderate habitats and give insights into mechanisms of cold adaptation. We propose provisional classification of the novel nitrite oxidizing bacterium as ‘Candidatus Nitrotoga arctica’.
Similar content being viewed by others
Introduction
Nitrification – the biological oxidation of ammonia to nitrite and further to nitrate is of fundamental importance for the global nitrogen cycle in marine and terrestrial habitats. It is a two-step, aerobic process performed by ammonia and nitrite oxidizing bacteria. Nitrite has a central position in the nitrogen cycle, connecting aerobic and anaerobic pathways. Nitrite oxidizing bacteria play a major role in removing nitrite from the environment because it is toxic for living organisms (Philips et al., 2002). If the linkage of nitrification and denitrification is disturbed, elevated nitrite concentrations may accumulate. At low pH, chemodenitrification is enhanced, which might stimulate N2O emission to the atmosphere.
Traditionally, classification of nitrifying bacteria is based on cell shape and ultrastructural criteria (Watson et al., 1989). Nitrite oxidizers catalyzing the second step of nitrification are so far restricted to four genera with eight described species. They are phylogenetically affiliated to the Alpha-, Gamma-, provisionally Deltaproteobacteria and the deep-branching bacterial phylum Nitrospirae (Spieck and Bock, 2005). Nitrifying bacteria are fastidious and slow-growing organisms, but their cultivation and isolation is indispensable to assign functions with sequence data obtained by direct molecular methods.
Temperature is one of the relevant factors influencing nitrification in nature with regard to activity as well as community structure (Avrahami and Conrad, 2003). Most of the described ammonia and nitrite oxidizing bacteria have a temperature optimum of about 28°C (Watson et al., 1989), increasing to 39°C in the case of Nitrospira moscoviensis (Ehrich et al., 1995). The temperature range for growth of nitrifying bacteria varies from −5°C (Jones and Morita, 1985) to about 60°C (Lebedeva et al., 2005).
The finding of marine crenarchaeota with the capability to oxidize ammonia (Könneke et al., 2005) indicated recently that nitrifying organisms are more diverse than previously assumed. Potential sources for novel nitrifying bacteria are extreme habitats like permafrost-affected soils, which are characterized by a low temperature mean and extreme temperature regimes (+25°C to −45°C) during seasonal freezing and thawing (Wagner et al., 2005). Little is known about the distribution of nitrifying bacteria in this environment (Lebedeva and Soina, 1994; Wagner et al., 2001). Cultivation approaches performed at 28°C with 3 mM substrate revealed the coexistence of Nitrobacter and Nitrospira in a soil sample derived from the rim of a low-centered polygon on Samoylov Island, Lena Delta, in a depth of 8–11 cm (Bartosch et al., 2002). In this study, enrichment of nitrite oxidizing bacteria was performed at low temperatures in accordance with the in situ conditions. Maximum soil temperature of the active layer in a depth of 7 cm on Samoylov Island amounted to 11.4°C in August 2001 (Kobabe et al., 2004).
Materials and methods
Investigation site
The study site is located in the youngest part of the Lena Delta on Samoylov Island (N 72°22, E 126°28). In this zone of continuous permafrost, a mean annual air temperature of –11.9°C was measured between 2001 and 2003 (Liebner and Wagner, 2007). The island is characterized by a pattern of low-centered ice-wedge polygons. Typical C/N values for the polygon rim are 16–18 and 31–37 for the wet polygon center (Kobabe et al., 2004). A total of nine samples was taken in August 2001 in a depth of 0–5 cm as a transect from the rim to the center (6675–6683) with a distance of 50 cm. Soil material was transported to Germany in a frozen state.
Culturing
Initially, 1 g of soil material was inoculated into 10 ml of mineral medium for nitrite oxidizing bacteria (Ehrich et al., 1995) with a substrate concentration of 0.3 mM. For plating, the lithoautotrophic medium with 3 mM nitrite and agarose (15 g l−1) was used. Parallel incubations were performed at 4°C, 10°C and 17°C. Consumption of nitrite was regularly checked by the Griess–Ilosavay spot test and replaced in the case of growth. Upscaling was performed as described (Spieck et al., 2006) to get a sufficient cell amount for further analysis. Purity tests were performed and modified by Steinmüller and Bock (1976).
Chemical analyses
Nitrite and nitrate concentrations were determined by ion-pair chromatography with a Hypersil ODS C18 column (125 × 4.6 mm) (Meincke et al., 1992) followed by UV detection in an automated system (Kontron, Eching, Germany).
Electron microscopy
For observation by electron microscopy cells were collected at 13 000 r.p.m., fixed with 2.5% (v/v), glutaraldehyde and 2% (w/v) osmium tetroxide and embedded in Epon 812 (Serva) according to a previously published protocol (Spieck et al., 2006). Examination was carried out with a transmission electron microscope (Zeiss model Leo 906E with a CCD camera model 794).
Fluorescence in situ hybridization and immunofluorescence labeling
Cells were collected and fixed in 3% paraformaldehyde as described by Bartosch et al. (1999). Fluorescent in situ hybridization (FISH) was performed according to the same protocol with the oligonucleotide probe Bet 42a (Manz et al., 1992). For identification of the novel betaproteobacterium, hybridization was carried out with the Cy3-labeled oligonucleotide NTG840 (see below) using a formamide concentration of 10–20%.
The nitrite oxidizing cells were detected with antibodies against their key enzyme by immunofluorescence (IF) labeling. Harvested cells were incubated with the antibodies Hyb 153-3 and Cy3-labeled secondary antibodies (Biotrend, Köln, Germany) (Bartosch et al., 1999). Control preparations without primary antibodies were included in every experiment. To detect total cells, samples were stained with 4′,6-diamidino-2-phenylindole (Dapi) at a concentration of 10 μg ml−1 for 5 min. Dapi staining was visualized by Leica filter set A (BP 340–380 exc; RKP 400; LP 425 em.). Cy3 labeling was visualized by Leica filter set N 2.1 (BP 515–560 exc; RKP 580; LP 590 em.).
Analysis of the fatty acids
Biomass for fatty acid extraction was obtained from large-scale enrichments grown in 1.5 l medium in 3 l Erlenmeyer flasks. Cultures were incubated without stirring for 2–6 months and regularly fed with sterile nitrite solution. When dense flocs of nitrite oxidizing bacteria had developed, cells were harvested by centrifugation, washed in 0.9% (w/v) NaCl and stored at −20°C. Preparation of fatty acid methyl esters and gas chromatographic analyses of the extracts were performed as described previously (Spieck et al., 2006).
16S rRNA gene sequence and phylogenetic analysis
The partial 16S rDNA fragments were amplified by PCR with the eubacterial primer set 341F/907R (Muyzer et al., 1998). As usual for TGGE/DGGE analysis, a GC-clamp was added to the forward primer. Denaturing gradient gel electrophoresis (DGGE) (Muyzer et al., 1998) was performed with a gradient from 50% to 80% denaturants and the temperature was 59°C. Bands were extracted from the gel, reamplified and the partial 16S rDNA sequences were compared with those on publicly accessible databases by using the program Basic Local Alignment Search Tool (BLAST, NCBI; Altschul et al., 1990). By the use of the program package ARB (Ludwig et al., 2004), a neighbor-joining tree was generated from an alignment of 16S rDNA sequences from selected Betaproteobacteria and the nitrite oxidizing enrichment culture 6680.
Cloning of the 16S rDNA
The 16S rRNA genes were amplified by the bacterial conserved primers 27F and 1492R. The PCR product was directly ligated into the pGEM-T vector cloning system (Promega, Mannheim, Germany) and transformed into competent cells as described in the manufacturer's instructions. For partial and near-complete sequencing of clone inserts, the plasmid primer SP6 and T7 were used to reamplify the insert. A range of bacterial conserved primers (341F/R, 517F/R, 907F/R, 1017F/R) were used for sequencing (Lane, 1991).
Specific PCR/specific oligonucleotide probe
On the basis of the newly retrieved 16S rDNA sequence of the novel betaproteobacterium (culture 6680), a new specific primer set and probe were constructed (NTG200F 5′-ctcgcgttttcggagcgg3′ and NTG840R 5′-ctaaggaagtctcctccc3′). The specificity was evaluated by using the program BLAST (NCBI) and Probe Match (RDP-II, Cole et al., 2007). The PCR conditions were as follows: step 1 96°C 2 min, step 2 58°C 50 s, step 3 72°C 50 s, step 4 final elongation 4 min. Steps 2 and 3 were repeated 28 times.
Results and discussion
Enrichment of the novel nitrite oxidizing bacterium
The investigation site, located on Samoylov Island in the Lena Delta, is characterized by a microrelief of ice-wedge polygons, with a depressed wet center and an elevated dryer rim (Kobabe et al., 2004). Selective enrichment of nitrite oxidizing bacteria was successfully performed from all nine soils of a polygon transect with incubation temperatures of 4°C, 10°C and 17°C using 0.3 mM nitrite. Following the protocol for the enrichment of a slow-growing Nitrospira strain (Spieck et al., 2006), permafrost cultures were regularly supplemented with additional substrate, when it was consumed. The highest degree of enrichment of nitrite oxidizing bacteria in relation to the total cell number was achieved in culture 6680, which was purified by plating on lithoautotrophic nitrite agar.
Physiological investigations of nitrite oxidizing enrichment cultures
To prove cold adaptation of the nitrite oxidizers obtained from permafrost-affected soils, growth of one enrichment culture (6678), originating from the polygon slope, was investigated in lithoautotrophic medium. Using 4°C for incubation, 0.9 mM nitrite was oxidized stoichiometrically to nitrate in about 1 month (not shown). In contrast, no growth was detected at an incubation temperature of 25°C. In control experiments at 4°C with different strains of Nitrobacter and Nitrospira 3 mM respectively 0.3 mM nitrite were not consumed within 2 months.
Chemotaxonomic analysis
Nitrite oxidizing bacteria are scattered in different subclasses of the phylogenetic tree, which is reflected in great variances of the fatty acid profiles (Lipski et al., 2001). Recently, chemotaxonomic analysis was successful in separating a new species of Nitrospira enriched from activated sludge (Spieck et al., 2006) from known species of this genus. Here, this method was used for the initial characterization of nitrite oxidizing enrichments originating from the Siberian Arctic. The fatty acid profile of the permafrost culture 6678 grown at 4°C, 10°C and 17°C was characterized by three hydroxy compounds (10:0 3OH, 12:0 3OH and 14:0 2OH) in combination with 16:0 and 16:1 cis 9 as major lipids (Table 1). This profile is clearly different from all nitrite oxidizing bacteria detected till now. In contrast to culture 6678, the long-chain fatty acid 18:1 cis 11 as dominating compound is typical for the genera Nitrobacter and Nitrococcus, whereas the marine genus Nitrospina can be differentiated by high amounts of the short-chain lipid 14:0. The genus Nitrospira including the Nitrospira dominated enrichment cultures Ns (42°C), Ns (47°C) and ‘Candidatus Nitrospira defluvii’ contain the fatty acid 16:1 cis 7 in combination with 16:1 cis 11 and/or 16:0 11methyl (Lipski et al., 2001; Spieck et al., 2006). These chemotaxonomic markers were almost absent in the enrichment culture 6678. Our results suggested the existence of one so far unknown low-temperature-adapted nitrite oxidizer. The adaptation is obviously realized by increasing the percentage of 16:1 cis 9 with decreasing temperature (Table 1), which is a strategy already described for ammonia oxidizing bacteria (Jones and Prahl, 1985) and bacterial isolates from permafrost soils (Ponder et al., 2005). Further enrichment cultures 6679, 6680 and 6681 incubated at 4°C, 10°C or 17°C resulted in this typical pattern for Betaproteobacteria with 16:0 and 16:1 cis 9 as dominating compounds, too (not shown). From these enrichments, only from culture 6675 (derived from the dryer polygon rim) grown at 17°C, a fatty acid pattern (16:0 and 16:1 cis 11) in the Nitrospira range was obtained (not shown), corresponding to that of ‘Candidatus Nitrospira defluvii’ (Spieck et al., 2006).
Identification of cold-adapted nitrite oxidizing bacteria with monoclonal antibodies
Results of fluorescence microscopic analysis using Dapi revealed that nearly all cells of the enrichment culture 6678 grown at 10°C appeared as irregular coccoid cells occurring in dense aggregates (Figure 1a). This morphology is not typical for one of the known genera in this bacterial group, but resembles the marine genus Nitrococcus. Functional identity of such cells as nitrite oxidizers was confirmed with monoclonal antibodies recognizing the key enzyme of nitrite oxidation (Aamand et al., 1996). The antibody Hyb 153-3 was shown to detect all known representatives of nitrite oxidizing bacteria specifically (Bartosch et al., 1999) and therefore offers a high potential for recognition of novel nitrite oxidizers. As shown in Figure 1a and b, clear reaction of the monoclonal antibodies with the coccoid cells became visible.
FISH
The results of fatty acid profiling shown above, revealed that most of the bacteria enriched in lithoautotrophic media with nitrite as energy source belong to Betaproteobacteria. To prove identity of the enriched coccoid cells, FISH was applied using the oligonucleotide probe Bet 42a targeting Betaproteobacteria, one of the major bacterial groups in Siberian tundra soils (Kobabe et al., 2004). Labeling was nearly complete in culture 6678 grown at 10°C where coccoid cells dominated (Figure 2a and b). At long-term incubation for 4 years at 17°C, the amount of the signal decreased drastically and only a few aggregates of coccoid cells were labeled (Figure 2c and d). Dominating rods, belonging to heterotrophic bacteria as shown by purity tests, were not labeled with probe Bet 42a.
Ultrastructure of the novel nitrite oxidizing bacteria
Nitrifying bacteria can be taxonomically classified on the genus level by their morphology and ultrastructure, for example, by the presence or absence of intracytoplasmic membranes. Striking differences in ultrastructure to the known genera of nitrite oxidizing bacteria became obvious when ultrathin sections of cultures 6678 and 6680 grown at 10°C were investigated by electron microscopy (Figure 3a). A longish cytoplasm was surrounded by an extraordinary wide periplasmic space at the longitudinal axis resulting in an irregular coccoid overall appearance. In cross–sections, cells gave the impression of ‘fried eggs’. Cytoplasmic and outer membrane were not in close contact at the longitudinal axis like in most other Gram-negative bacteria, but cells showed a balloon-like periplasmic space (Figure 3b). An unusual wide periplasmic space was already described for members of the genus Nitrospira (Watson et al., 1986), but the dimensions were more extended here. No intracytoplasmic membranes or carboxysomes were found. Cells had an overall diameter of 0.4–0.7 μm and a maximal length of 1.0 μm.
Electron micrographs of ultrathin sections of the nitrite oxidizing enrichment culture 6678 grown at 10°C. (a) Overview of the dominant cell type with an unique ultrastructure. Bar=500 nm. (b) A longish cytoplasm is surrounded by an extraordinary wide periplasmic space. Cy=cytoplasm, cm=cytoplasmic membrane, om=outer membrane, P=periplasm. Bar=200 nm.
Analysis of the nitrite oxidizing enrichments by DGGE profiles
To classify nitrite oxidizers in the enrichment cultures 6675, 6678, 6679 and 6680 grown at 10°C or 17°C, molecular studies were performed. DNA was extracted, the 16S rDNA fragments were amplified with the eubacterial primer set 341F/907R and separated by DGGE. In DNA extracted from culture 6680, only one clear band was obvious in the gel (Figure 4), although accompanying heterotrophic bacteria grow up in complex organic media. This band only occurred when the culture was grown chemolithoautotrophically (line 1) and was absent when the cells were supplied with organic substrates without addition of nitrite (line 2). Bands in the same range (lines 3–6) were found as dominating ones in DNA extracted from cultures 6675 (17°C), 6678 (17°C), 6679 (17°C) and 6678 (10°C). In contrast to culture 6678 cultivated for a few months at 17°C, the unique DNA band became very weak when the enrichment of the same soil sample was incubated at 17°C for 4 years (line 7). Simultaneously other bands indicating the presence of potential heterotrophic bacteria appeared. Accordingly, in electron microscopic investigations of culture 6678 grown at 17°C, the percentage of coccoid cells was reduced drastically and different cell types of bacteria, mostly in the form of rods, frequently occurred (not shown).
Denaturing gradient gel electrophoresis (DGGE) profile of nitrite oxidizing enrichment cultures derived from permafrost soil cultivated at 10°C or 17°C. Line 1: culture 6680 grown lithoautotrophically at 10°C; line 2: culture 6680 grown with organic material; line 3: culture 6675 (17°C); line 4: 6678 (short-term incubation at 17°C); line 5: 6679 (17°C); line 6: 6678 (10°C); line 7: 6678 (long-term incubation at 17°C); line 8: pure culture of Nitrospira moscoviensis M1; line 9: pure culture of Nitrobacter hamburgensis X14. Arrow represents Nitrospira-like bacterium.
Following long-term incubation of the soil material, exclusively one weak Nitrospira-like band (Figure 4, arrow) was detected and sequenced (enrichment 6675 at 17°C). Here, the occurrence of Nitrospira is in accordance with the detection of typical fatty acids for this genus mentioned above. However, DGGE analysis was performed 1 year later than chemotaxonomic analysis. As a consequence, the Nitrospira-like bacterium was overgrown by the typical betaproteobacterium (line 3).
16S rRNA gene sequence and phylogenetic analysis
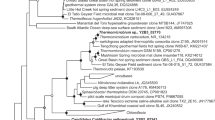
In DGGE analysis shown above, most nitrite oxidizing enrichments were dominated by the same DNA band. To clarify phylogeny of the bacterial source, bands belonging to the cultures 6678 and 6680 enriched at 10°C (Figure 4, lines 1+6) were extracted from the gel, reamplified and sequenced. Analysis of partial 16S rRNA gene sequences showed that both cultures were identical. They fell into a cluster of the Betaproteobacteria, which consisted exclusively of cloned environmental samples. TA cloning was performed with culture 6680 and the almost complete sequence was obtained by the use of a range of several bacterial conserved primers. The sequence has been deposited in GenBank under the accession number DQ839562. Phylogenetic relationship between cloned sequences from the culture 6680 and representatives of the class Betaproteobacteria is shown in Figure 5. For the novel organism the name ‘Candidatus Nitrotoga arctica’ is proposed.
Comparative sequence analysis of the 16S rRNA genes revealed a high level of sequence identity between the new nitrite oxidizing betaproteobacterium and clone Run-S67 (AB247475) from domestic sewage (99.0%), clone Elb 168 (AJ421928) from a biofilm of a polluted river (98.8%) (Brümmer et al., 2003), clone BG.g12 (DQ228379) from subglacial environments (97.8%) (Skidmore et al., 2005) and clone c5LKS43 (AM086129) from sediments of the mesotrophic lake Kinneret (96.3%) (Schwarz et al., 2007). The next related taxonomically described organism is Gallionella ferruginea (93.5% similarity).
Development of a specific PCR/specific oligonucleotide probe
To prove the correlation of the 16S rRNA sequence with the cells observed in light and electron microscopy, a specific oligonucleotide probe (NTG840) was designed. Using 10–20% formamide concentration, dominating coccoid cells following incubation at 10°C revealed a positive reaction (not shown). Additionally, a new specific primer set (NTG 200F/NTG 840R) for the novel betaproteobacterium was constructed and evaluated. Whereas the sequence of NTG200F is complementary to the sequences of the cultures 6678/6680 and the next three relatives (Figure 5), the primer/probe NTG840R also targets several environmental sequences inside the Betaproteobacteria. Analysis of natural samples (6676 and 6678) as well as 10°C enrichments of samples 6675, 6678, 6679, 6680, 6681 and 6682 revealed that the new betaproteobacterium is detectable by this specific primer set (not shown).
Ecological aspects
Phylogenetic analyses of permafrost soils indicated that the majority of the clone sequences were 5–15% different from those in the current databases (Zhou et al., 1997). The results presented here enabled identification of one of the unknown genera and gave insight into its physiology and function, which remained unknown for most of the bacteria active in situ. On the basis of the first characteristics presented here, existence of an additional genus of nitrite oxidizing bacteria in polygonal tundra soil in Siberia is indicated. In contrast to terrestrial members of Nitrobacter and Nitrospira, cells of these Betaproteobacteria are well adapted to low temperatures. Our data suggest that this organism is active at temperatures up to 22°C and at very low nitrite concentrations (0.3 mM) in permafrost-affected soils. The finding of a so far unkown nitrite oxidizing bacterium is also important for moderate habitats like waste water treatment plants. Transfer of the preferred culture conditions to activated sludge in Dradenau, Hamburg revealed that the same organism is present, but has not been recognized before. Here, bacteria with nearly identical 16S rRNA sequence and ultrastructure were obtained when selective enrichment of nitrite oxidizing bacteria was performed at 10°C and not as usual at 28°C. The ongoing activity of nitrifying bacteria at low temperature with simultaneous inhibition of heterotrophic bacteria (including denitrifiers) is of high relevance for biotechnology.
Taxonomical considerations
Based on the results of this study, we propose, according to Murray and Stackebrandt (1995), provisional classification of the novel nitrite oxidizing bacterium as ‘Candidatus Nitrotoga arctica’. The short description of ‘Candidatus Nitrotoga arctica’ gen. nov, sp. nov is as follows:
Ni.tro.to’ga L.n. nitrum nitrate; L. fem.n. toga Roman outer garment. Arc.ti.ca named after the place where the organism was first discovered.
Coccoid cells to short rod-shaped cells with a size of 0.4–0.7 × 1.0 μm. Periplasmic space is extraordinary wide, irregular bollooning over the longitudinal axis. Intracytoplasmic membranes and carboxysomes are absent. Gram-negative, non-motile. Multiplication by binary fission. Cells aggregate to flocs. Aerobic chemolithotroph metabolism with oxidation of nitrite to nitrate. Carbon dioxide is used as the sole carbon source. Non-marine. Growth range between 4°C and 22°C with an optimum at 10°C. Adapted to low nitrite concentrations (0.3 mM) with a tolerance limit of 1.2 mM.
Dominating fatty acids are 16:1 cis 9 and 16:0. Hydroxy fatty acids 10:0 3OH, 12:0 3OH and 14:0 2OH are present. Phylogenetically assigned to the Betaproteobacteria based on 16S rRNA gene sequences (accession no. DQ839562). Not isolated.
Habitat: active layer of permafrost-affected soils in the Siberian Arctic. Cells with nearly identical ultrastructure and 16S rRNA gene sequence were also detected in activated sludge from a municipal waste water treatment plant in Hamburg, Germany.
Perspective
Isolation of the novel nitrite oxidizing organism is in progress, which will enable investigations of physiological aspects like organotrophic growth and the capability for nitrate reduction. Correlation of structure and function is another main point with regard to the conspicuous ultrastructure. Molecular and biological studies will focus on the detection in environmental samples and distribution patterns of the different genera of nitrite oxidizing bacteria.
Accession codes
References
Aamand J, Ahl T, Spieck E . (1996). Monoclonal antibodies recognizing nitrite oxidoreductase of Nitrobacter hamburgensis, N. winogradskyi, and N. vulgaris. Appl Environ Microbiol 62: 2352–2355.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ . (1990). Basic local alignment search tool. J Mol Biol 215: 403–410.
Avrahami S, Conrad R . (2003). Patterns of community change among ammonia oxidizers in meadow soils upon long-term incubation at different temperatures. Appl Environ Microbiol 69: 6152–6164.
Bartosch S, Hartwig C, Spieck E, Bock E . (2002). Immunological detection of Nitrospira-like bacteria in various soils. Microb Ecol 43: 26–33.
Bartosch S, Wolgast I, Spieck E, Bock E . (1999). Identification of nitrite oxidizing bacteria with monoclonal antibodies recognizing the nitrite oxidoreductase. Appl Environ Microbiol 65: 4126–4133.
Brümmer IHM, Felske A, Wagner-Döbler I . (2003). Diversity and seasonal variability of β-Proteobacteria in Biofilms of polluted rivers: analysis by temperature gradient gel electrophoresis and cloning. Appl Environ Microbiol 69: 4463–4473.
Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM et al. (2007). The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res 35 (database issue): D169–D172; doi:10.1093/nar/gkl889.
Ehrich S, Behrens D, Lebedeva E, Ludwig W, Bock E . (1995). A new obligately chemolithoautotrophic, nitrite oxidizing bacterium, Nitrospira moscoviensis sp: nov and its phylogenetic relationship. Arch Microbiol 164: 16–23.
Jones RD, Morita R . (1985). Low-temperature growth and whole cell kinetics of a marine ammonium oxidizer. Mar Ecol Prog Ser 21: 239–243.
Jones RD, Prahl FG . (1985). Lipid composition of a marine ammonium oxidizer grown at 5 and 25°C. Mar Ecol Prog Ser 26: 157–159.
Kobabe S, Wagner D, Pfeiffer E-M . (2004). Characterization of microbial composition of Siberian tundra soil by fluorescence in situ hybridization. FEMS Microbiol Ecol 50: 13–23.
Könneke M, Bernhard AE, De La Torre JR, Walker CB, Waterbury J, Stahl DA . (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437: 543–546.
Lane DJ . (1991). 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds). Nucleic Acid Techniques in Bacterial Systematics. Academic Press: Chichester, UK, pp 115–175.
Lebedeva EV, Alawi M, Fiencke C, Namsaraev B, Bock E, Spieck E . (2005). Moderately thermophilic nitrifying bacteria from a hot spring of the Baikal rift zone. FEMS Microbiol Ecol 54: 297–306.
Lebedeva E, Soina V . (1994). Nitrifying bacteria as a promising group for paleoecological investigations in the permafrost. In: Gilichinsky DA (ed). Viable Microorganisms in Permafrost. First International Conference on Cryopedology and Global Change. Russian Academy of Sciences Pushino Research Centre, ISBN 5-206-10586-6, pp 74–82.
Liebner S, Wagner D . (2007). Abundance, distribution and potential activity of methane oxidizing bacteria in permafrost soils from the Lena Delta, Siberia. Environ Microbiol 9: 107–117.
Lipski A, Spieck E, Makolla A, Altendorf K . (2001). Fatty acid profiles of nitrite-oxidizing bacteria reflect their phylogenetic heterogeneity. Syst Appl Microbiol 24: 377–384.
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhu K et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371.
Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H . (1992). Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol 15: 593–600.
Meincke M, Bock E, Kastrau D, Kroneck PMH . (1992). Nitrite oxidoreductase from Nitrobacter hamburgensis: redox centers and their catalytic role. Arch Microbiol 158: 127–131.
Murray RG, Stackebrandt E . (1995). Taxonomic note: implementation of the provisional status Candidatus for incompletely described procaryotes. Int J Syst Bacteriol 45: 186–187.
Muyzer G, Brinkhoff T, Nübel U, Santegoeds C, Schäfer H, Wawer C . (1998). Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. Mol Microbial Ecol Manual 3.4.4: 1–27.
Philips S, Laanbroek HJ, Verstraete W . (2002). Origin, causes, and effects of increased nitrite concentrations in aquatic environments. Rev Environ Sci Biotechnol 1: 115–141.
Ponder M, Gilmour SJ, Bergholz PW, Mindock CA, Hollinsworth R, Thomashow et al. (2005). Characterization of potential stress responses in ancient Siberian permafrost psychroactive bacteria. FEMS Microbiol Ecol 53: 103–115.
Schwarz JI, Eckert W, Conrad R . (2007). Community structure of Archaea and Bacteria in a profundal lake sediment Lake Kinneret (Israel). Syst Appl Microbiol 30: 239–254.
Skidmore M, Anderson SP, Sharp M, Foght J, Lanoil BD . (2005). Comparison of microbial community compositions of two subglacial environments reveals a possible role for microbes in chemical weathering processes. Appl Environ Microbiol 71: 6986–6997.
Spieck E, Bock E . (2005). The lithoautotrophic nitrite-oxidizing bacteria. In: Garrity G, Brenner DJ, Krieg NR, Staley JT (eds). Bergey's Manual of Systematic Bacteriology, 2nd edn. Springer-Verlag: New York, NY, pp 149–153.
Spieck E, Hartwig C, McCormack I, Maixner F, Wagner M, Lipski A et al. (2006). Selective enrichment and molecular characterisation of a previously uncultured Nitrospira-like bacterium from activated sludge. Environ Microbiol 8: 405–415.
Steinmüller W, Bock E . (1976). Growth of Nitrobacter in the presence of organic matter; I. Mixotrophic growth. Arch Microbiol 108: 299–304.
Wagner D, Lipski A, Embacher A, Gattinger A . (2005). Methane fluxes in permafrost habitats of the Lena Delta: effects of microbial community structure and organic matter quality. Environ Microbiol 7: 1582–1592.
Wagner D, Spieck E, Pfeiffer E-M, Bock E . (2001). Microbial life in terrestrial permafrost: Methanogenesis and nitrification in gelisols as potentials for exobiological processes. In: Horneck G, Baumstark-Khan C (eds.) Astrobiologie-the Quest for the Conditions of Life. Springer-Verlag: Berlin, NY, pp 143–159.
Watson SW, Bock E, Valois FW, Waterbury JB, Schlosser U . (1986). Nitrospira marina gen. nov. sp. nov: a chemolithotrophic nitrite-oxidizing bacterium. Arch Microbiol 144: 1–7.
Watson SW, Bock E, Harms H, Koops H-P, Hooper AB . (1989). Nitrifying bacteria. In: Staley JT, Bryant MP, Pfennig N, Holt JG (eds). Bergey's Manual of Systematic Bacteriology, 1st edn, Vol. 3. The Williams & Wilkins Co: Baltimore, MD, pp 1808–1834.
Zhou J, Davey ME, Figuera JB, Rivkina E, Gilichinsky D, Tiedje JM . (1997). Phylogenetic diversity of a bacterial community determined from Siberian tundra soil DNA. Microbiology 143: 3913–3919.
Acknowledgements
Sampling was kindly performed by D Wagner (AWI Potsdam) in frame of the German–Russian cooperation ‘System Laptev Sea’. We thank I Wachholz for technical help in electron microscopy, P-G Jozsa for chemical analysis by HPLC-technique and M Kaya for culturing nitrite oxidizers from activated sludge. FISH analysis was kindly supported by C Hartwig. Laboratory assistance was provided by M Klimova (Voronezh State University) in the frame of a DAAD exchange programe. This research was funded by the BMBF Russian–German Cooperation ‘Laptev Sea System: Process Studies on Permafrost Dynamics in the Laptev Sea’.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Alawi, M., Lipski, A., Sanders, T. et al. Cultivation of a novel cold-adapted nitrite oxidizing betaproteobacterium from the Siberian Arctic. ISME J 1, 256–264 (2007). https://doi.org/10.1038/ismej.2007.34
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2007.34
Keywords
This article is cited by
-
Diversity and distribution of iron-oxidising bacteria belonging to Gallionellaceae in different sites of a hydroelectric power plant
Brazilian Journal of Microbiology (2024)
-
Relevance of Candidatus Nitrotoga for nitrite oxidation in technical nitrogen removal systems
Applied Microbiology and Biotechnology (2021)
-
Shifts in the Abundance and Community Composition of Particle-Associated and Free-Living Nitrospira Across Physicochemical Gradients in the Pearl River Estuary
Estuaries and Coasts (2021)
-
The Nitrogen-Cycling Network of Bacterial Symbionts in the Sponge Spheciospongia vesparium
Journal of Ocean University of China (2021)
-
Extremophilic nitrite-oxidizing Chloroflexi from Yellowstone hot springs
The ISME Journal (2020)