Abstract
The afro-alpine region comprises the high mountains of Ethiopia and tropical East Africa, which represent biological ‘sky islands’ with high level of endemism. However, some primarily arctic-alpine plants also occur in the afro-alpine mountains. It has been suggested that these plants are Tertiary relicts, but a recent worldwide study of Arabis alpina suggests that this species colonized the region twice during the Pleistocene. Here we investigate the detailed colonization history of A. alpina in the afro-alpine region based on chloroplast DNA sequences from 11 mountain systems. The results confirm the twice-into-Africa scenario. The Asian lineage is confined to the mountains closest to the Arabian Peninsula, on opposite sides of the Rift Valley (Simen Mts and Gara Muleta in Ethiopia), suggesting long-distance dispersal of this lineage. The African lineage is divided into two phylogeographic groups with distinct geographic distribution. The observed pattern is consistent with isolation of the African lineage in at least two interglacial refugia, located on separated highlands, followed by range expansion in cooler period(s), when the afro-alpine habitat extended further down the mountains. Several long-distance dispersal events, also across the Rift Valley, are suggested by single haplotypes observed outside the area occupied by the phylogeographic groups they belonged to.
Similar content being viewed by others
Introduction
The afro-alpine region encompasses the high mountains of Ethiopia and tropical East Africa (Kenya, Tanzania and Uganda; Hedberg, 1969, 1970). Most mountains in this region are of volcanic origin. Whereas the Ethiopian mountains are part of quite extensive highlands, the southern part of the region consists mainly of isolated peaks emerging from the lowlands and the elevated shoulders of the Great Rift Valley. Along the slopes of these mountains, the vegetation is divided into three altitudinal zones (Hedberg, 1951): the afro-montane zone, the sub-alpine ericaceous zone and, above 3500 m, the afro-alpine zone. Endemism is high in the afro-alpine flora (80% at the species level), indicating that this flora has long been isolated from other high mountain or temperate floras (Hedberg, 1969). However, a Northern Hemisphere temperate element makes up as much as 13% of the afro-alpine plants. The time and source of immigration for these plants has been a subject of debate. While some authors considered them to be Tertiary relicts (Hedberg, 1969, 1970), more recent immigration has also been suggested (Koch et al., 2001, 2006).
Another interesting question is how the different ‘sky islands’ were colonized and to what degree plant populations on these mountains have remained isolated. Long-distance dispersal by wind, notably by cyclones, and/or by birds from one mountain to another has been proposed as mechanisms for plant dispersal within the fragmented afro-alpine region (Hedberg, 1969). However, it is also likely that the Pleistocene climate changes have played an important role for the dispersal among mountain tops. During glaciations in the northern Hemisphere, the African tropics were cooler and drier (Bonnefille et al., 1990; deMenocal, 1995). While the top of many mountains was covered by glaciers, both the afro-alpine and the ericaceous zone extended about 1000–1500 m lower than today and thus occupied considerably larger areas (Flenley, 1979; Gottelli et al., 2004). Thus, during the last glaciation, the Ethiopian highlands on both sides of the Great Rift Valley (Figure 1) were covered by afro-alpine heaths and grassland (Messerli et al., 1977), and provided habitat for a large population of the endemic Ethiopian wolf, a specialized predator of the afro-alpine zone (Gottelli et al., 2004). Also in the mountain massifs of tropical East Africa, of which some are more isolated (for example, Mt Kilimanjaro and Mt Meru), afro-alpine populations may have been larger and distances among habitat patches smaller during the cool periods of the Pleistocene, facilitating dispersal from one mountain to another (Moreau, 1963; Hedberg, 1986; Livingstone, 1993).
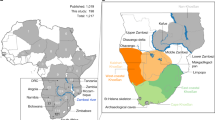
Map of the study area and cpDNA haplotype network. For each mountain massif, a pie diagram shows the observed haplotypes. The colours in the diagram correspond to the colours of the haplotypes in the network. The area shaded in grey indicates mountains and highlands. The thick dark-grey lines show the Great Rift Valley. On the haplotype network, the three different lineages are indicated. The size of the haplotypes is drawn proportional to their frequencies. Thick lines represent substitutions, thin lines insertions or deletions and small circles hypothetical intermediate haplotypes. The European lineage is represented in a simplified way and haplotypes not observed in Africa are plotted in white.
Migration into the afro-alpine region from the north may also have been facilitated during cooler periods of the Pleistocene. The mountains bordering the Red Sea may have formed stepping stones for alpine plants, connecting Eastern Europe and the Caucasus through the Middle East to northeast Africa (Kingdon, 1990). Gottelli et al. (2004) suggested that the Ethiopian wolf evolved from a Eurasian grey wolf-like ancestor which followed this route during the late Pleistocene, probably as late as during the last glacial period (∼100 000 years ago).
Arabis alpina L. (Brassicaceae) is both an afro-alpine and arctic-alpine plant with a wide distribution range, extending from the northern amphi-Atlantic region and the European mountains including the Mediterranean to the Caucasus, and to the East African mountain regions over the Arabian Peninsula (Koch et al., 2006). It is a perennial rosette plant with seeds that lack typical mechanisms promoting long-distance dispersal. In Ethiopia and tropical East Africa, A. alpina occurs in the afro-alpine zone above 3500 m as well as in the ericaceous zone (3000–4000 m), in moist habitats with open gravel or rocks (Hedberg 1962, 1986).
In a recent study of the range-wide phylogeography of A. alpina based on sequences of chloroplast DNA (cpDNA) and the internal transcribed spacers of ribosomal DNA (ITS), Koch et al. (2006) inferred that the species most likely originated in the Middle East. From there, a European lineage spread westwards and northwards to give rise to the European and arctic populations, and an African lineage spread to the eastern African mountains. A third lineage established in the Middle East, and expanded to the Caucasus in the east and through Yemen to the Ethiopian mountains (the Asian lineage). Koch et al. (2006) thus concluded that the mountains in East Africa were colonized by two independent lineages. They dated the divergence between these two lineages to approximately 500 000 years ago, rejecting Hedberg's (1969, 1970) hypothesis that the African plants are Tertiary relicts. The estimated date is based on a mutation rate for Brassicaceae estimated from the divergence between Cardamine and Barbarea, which was considered to predate the Pliocene and to be about 6 mya old (Koch et al., 2000, 2001; dating based on the occurrence of Rorippa pollen, a close relative of Cardamine and Barbarea, in geological samples being 2.5–5 mya old and dating back to the Pliocene, cited by Mai, 1995). Koch et al's. (2006) study included, however, only seven plants from Africa and could thus neither address the distribution of the two lineages nor the history of colonization of the different mountain massifs.
Here we use samples from 11 different mountain massifs in eastern Africa to investigate the distribution of the two divergent cpDNA lineages of African A. alpina. We analyse the phylogeographic pattern within the afro-alpine region to gain a better understanding of the history of these isolated populations.
Materials and methods
Plant material
A. alpina was collected in 2003 and 2004 from 11 mountain regions: Simen Mts, Bale Mts, Mt Choke, Mt Kaka, Mt Chilallo and Gara Muleta in Ethiopia; Mt Kenya, Aberdares and Mt Elgon in Kenya; and Mt Kilimanjaro and Mt Meru in Tanzania (Figure 1). On each mountain, if possible, we collected plants from three distinct populations separated by one to more than 100 km. Whenever possible, samples were collected at about 10 m distance. Fresh leaves were dried in silica gel. CpDNA was sequenced from six individuals from each mountain, two from each sampled population (66 individuals in total, Appendix A).
Molecular methods
Genomic DNA was extracted using the DNeasy extraction kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. As Koch et al. (2006), we amplified the trnL intron and trnL-trnF intergenic spacer (IGS). We used the universal primers c (5′-CGAAATCGGTAGACGCTACG-3′) and d (5′-GGGGATAGAGGGACTTGAAC-3′; Taberlet et al., 1991), which anneal in the first and second exon of the trnL gene, and e (5′-GGTTCAAGTCCCTCTATCCC-3′; Taberlet et al., 1991) and the primer 5′-GATTTTCAGTCCTCTGCTCTAC-3′ (Dobeš et al., 2004), which anneal in the second exon of the trnL and the trnF genes. PCR reactions were performed in a total volume of 10 μl containing 1 × PCR buffer, 3 mM MgCl2, 0.8 mM dNTP (total concentration; Applied Biosystems, Foster City, CA, USA), 0.4 μl of each primer (10 μ M), 0.5 U AmpliTaq polymerase (Perkin-Elmer, Waltham, MA, USA) and 2 μl template DNA (diluted 1:10). Amplifications were performed using a PTC-200 Thermal Cycler (MJ Research, Waltham, MA, USA). The PCR programme consisted of 3 min at 95°C followed by 35 cycles each comprising 30 s at 95°C, 30 s annealing at 57°C for trnL intron and at 55°C for trnL-trnF IGS, and 60 s at 72°C, followed by a final extension for 7 min at 72°C. PCR products were checked on 1% agarose gels, purified with exonuclease I and shrimp alkaline phosphatase (ExoSAP) and sequenced using the TaqDyeDeoxy Terminator Cycle Sequencing Kit (Applied Biosystems). The original amplification primers were used, except for the reverse trnL/F IGS primer, which was modified with an extra C at the 3′-end (Dobeš et al., 2004). Electrophoresis was carried out on an ABI 3100 capillary sequencer (Applied Biosystems). Both strands were sequenced, and in most cases the complete sequence could be read in both directions.
Data analysis
The sequences were edited and manually aligned using GeneTool 2.0. (BTI Software, Edmonton, Canada). We combined our data with the data set of Koch et al. (2006), comprising haplotypes from the whole range of the species, to see how our data fit into the previously published phylogeny. A haplotype network was plotted using the software Network 4.112 (Bandelt et al., 1999). Among the African sequences, the only gaps were observed in a stretch of several Cs (3–6) followed by several Ts (9–12). We assumed that the mutation rate for length variation in this region was higher than for substitutions, and thus that homoplasy was more likely for these characters. Therefore, the African part of the network was plotted by first excluding gaps and then manually adding haplotypes differing by the number of Cs and Ts, considering these length variants of the haplotypes defined by the nucleotide sequences. A maximum parsimony tree was constructed using the software TNT (Goloboff, 1999). We performed a traditional search, starting with 10 random addition sequences and using TBR branch swapping. Gaps composed of several base pairs were recoded as single characters, as were gaps consisting of single base pair steps. Insertions in the poly-C and poly-T stretch were weighed down to 0.5. Sequences of Aubrieta deltoidea and Draba aizoides were used as outgroups (obtained from Koch et al., 2006). A strict consensus tree was computed and support for the different clades was estimated from 1000 bootstrap replicates. We also plotted a neighbour-joining tree using Jukes and Cantor's (1969) distance and the program TREECON ver. 1.3b (Van de Peer and De Wachter, 1994). Gaps were coded as for the maximum parsimony analysis, but all weighed equally, and support assessed with 1000 bootstrap replicates. Furthermore, the phylogeny was inferred by a Bayesian approach using the software MrBayes (Ronquist and Huelsenbeck, 2003). The data were divided in two distinct partitions: the first consisted of the recoded gaps and the second of the DNA sequences. Evolutionary rates as well as rate variation parameters were estimated for each partition separately. For substitutions, a simple model with one substitution rate was assumed. The Markov Chain Monte–Carlo simulations were run for 4 million generations and samples were collected every 100 generations.
For the phylogeographic analyses, we included the seven African individuals from Koch et al. (2006). Genetic diversity was estimated for each of the 11 mountain regions as haplotype diversity (h) and nucleotide diversity (π; Nei, 1987). We quantified the level of genetic differentiation among phylogeographic groups and among mountains within groups by analysis of molecular variance (AMOVA) using the program Arlequin 3.01 (Excoffier et al., 2005). Because the phylogeographic pattern was clear on the map, we considered it superfluous to test alternative groupings. Significance of differentiation at the different levels was estimated from 10 000 permutations. Both FST, an estimate of differentiation based on haplotype frequencies only, and ΦST, an estimate taking into account the molecular distance between haplotypes (number of pair-wise differences), were estimated. These two estimators are analogous to GST and NST, respectively (Pons and Petit, 1996). In case of correspondence between haplotype phylogenies and their geographic distribution, estimates for NST (ΦST) will be greater than the GST (FST) values (Pons and Petit, 1996; Petit et al., 2005). A permutation test in the program PERMUT (available from http://www.pierroton.inra.fr/genetics/labo/Software) was used to test whether the difference between the two estimates was significant (Burban et al., 1999). As an alternative approach to investigate the statistical association between genealogy and geography, a phylogeographic nested clade analysis (NCPA, Templeton, 2004) was performed. We constructed a simplified network including only haplotypes occurring in Africa and grouped the haplotypes into nested clades according to the algorithm described by Templeton et al. (1987). This nested design, together with the geographical locations of the samples (coordinates), was analysed by means of permutation tests at the different nesting levels with the software GeoDis (Posada et al., 2000). Templeton's inference key (Templeton, 2004) was used to infer the processes likely to be involved in the statistically significant associations observed.
To investigate the demographic history of the populations, the mismatch distribution (the distribution of the number of pair-wise differences between haplotypes) was estimated for the African lineage. This distribution is usually multimodal in samples drawn from populations at demographic equilibrium, but unimodal in populations having experienced a recent demographic expansion (Rogers and Harpending, 1992). Assuming a model of sudden expansion under which the population size increased from N0 to N1 t generations ago, but was constant both before and after the expansion, it is possible to estimate the parameter τ=2 μt from the mismatch distribution, where μ is the mutation rate per DNA fragment. The software Arlequin 3.01 estimates this parameter by a generalized nonlinear least-square approach, according to Schneider and Excoffier (1999). The validity of the step-wise expansion model is tested by a parametric bootstrap approach, which is also used to estimate confidence intervals (CIs) for the estimated parameters.
Results
Haplotype relationships and geographic distribution
The length of the aligned cpDNA sequences from the 66 African individuals in our material was 745 bp. There were 14 variable sites, comprising eight substitutions and six indels. All indels were in the above-mentioned stretch of several Cs (3–6) followed by several Ts (9–12). Seven of the substitutions and all length variation were parsimony informative. Nine different haplotypes were detected in our material (Figure 1, Appendix A). Two of them, haplotypes 4 and 19, were identical to haplotypes observed by Koch et al. (2006) and the remaining seven were new (Koch et al. found also two additional African haplotypes, haplotypes 5 and 18, which were not observed in our material; Figure 1). The sequences of the new haplotypes have been deposited in GenBank (accession numbers EF449508–EF449514). Combining our data with those of the range-wide analysis of Koch et al. resulted in an alignment with 26 variable sites and 25 indels (excluding outgroups). Nine substitutions and 11 indels were parsimony informative in this combined data set.
The haplotype network of the combined data set confirmed the general structure reported by Koch et al. (2006). There were three major groups, the African, the Asian and the European group (Figure 1). Our new haplotypes were placed mainly in the African group (A, B, C, D, E and F), increasing its diversity, and one haplotype was placed in the Asian group (G). The maximum parsimony analysis using the combined matrix yielded 22 most parsimonious trees with a length of 215 steps. On the consensus tree, the African and the Asian haplotypes formed distinct clades with moderate bootstrap support (61 and 68%, respectively; Figure 2). Haplotype 17 from an individual sampled in Lebanon by Koch et al. (2006) was placed in a sister position to the African clade, but the bootstrap support was only 50%. Within the African clade, haplotypes A, B, C, D and F formed a sub-clade (bootstrap support 51%). The topography of the neighbour-joining tree and the Bayesian tree was nearly identical to the maximum parsimony tree. The Asian and the African clade, as well as the sub-clade containing haplotype A were statistically supported by these analyses as well (Figure 2). In addition, a second weakly supported sub-clade in Africa joining haplotypes 5, 18 and E was suggested (56% bootstrap support on the NJ-tree, 0.73 posterior probability on the Bayesian tree; not shown). In order to get a robust estimate for our shallow phylogeny, we also estimated a maximum parsimony tree excluding insertions and deletions. The main topography was still identical, but bootstrap support was 65% for the African clade and 60% for the sub-clade containing haplotype A (tree not shown).
Strict consensus tree of the 22 most parsimonious trees found in analysis of the cpDNA haplotypes of Arabis alpina. Three different estimates of the support for the nodes are given: bootstrap value from the maximum parsimony analysis and the neighbour-joining analysis (1000 replicates) and Bayesian posterior probability. The three different lineages are shown.
The Asian lineage was restricted to the northernmost and easternmost mountain regions in Ethiopia, situated on opposite sides of the Great Rift Valley (Simen Mts and Gara Muleta, Figure 1). Haplotype G, a derived haplotype of the Asian lineage, occurred only in Gara Muleta. In the Simen Mts, two haplotypes from the African lineage were found as well.
The haplotypes of the African lineage could be divided into two groups with largely non-overlapping geographic distributions, corresponding to the division observed in the network and the phylogenetic analyses (Figures 1 and 2). One of these phylogeographic groups consisted of the weakly supported clade containing haplotype A and will hereafter be referred to as group A (the common haplotype A and its derived haplotypes B, C, D and F; Figure 1.) The other contained the rest of the African haplotypes and will hereafter be referred to as group 19. Despite the fact that this group was not resolved and did not constitute a clade, its topology on the haplotype network and its distinct geographic distribution support it as a phylogeographic group. Group A was found mostly east of the Rift Valley, whereas group 19 occurred west of it. In the south of the study area, however, group 19 extended eastwards across the Rift Valley to Mt Meru and Mt Kilimanjaro (Figure 1). Haplotype 5 found on Mt Kilimanjaro occurred also once on Mt Kenya where it co-occurred with haplotypes of group A, extending group 19 further northwards. In the north, group A extended westwards across the Rift Valley with the derived haplotype C (found on Mt Kaka and in the Simen Mts) and overlapped with group 19 in the Simen Mts (where it also co-occurred with the Asian lineage). The Simen Mts were the only locality where haplotypes of all three phylogeographic groups co-occurred.
Phylogeography and demographic history
Genetic diversity estimates for each mountain region are presented in Table 1. Nucleotide diversity was highest in the Simen Mts with π=0.59%. Among the remaining sites, nucleotide and haplotype diversities were highest on Mt Kaka with π=0.36% and h=0.73. On three mountains, only a single haplotype was found (Mt Choke, Mt Chilallo and Mt Elgon).
For the AMOVA analyses, we divided the populations into three phylogeographic regions according to the geographic distributions of the Asian lineage (Simen in the north and Gara Muleta in the east) and of the two African haplotype groups (group A: Mt Chilallo, Mt Kaka, Bale Mts, Mt Kenya and Aberdares; group 19: Mt Choke, Mt Elgon, Mt Meru and Mt Kilimanjaro; Figure 1). High differentiation was observed among these three regions. The percentage of variation among regions was considerably higher when taking into account molecular variation (71.5%) than when analysing only haplotype frequencies (51.2%; Table 2). According to the GST/NST test, this difference was significant with P=0.014, confirming that at this scale, related haplotypes were grouped geographically. The differentiation between the two groups belonging to the African lineage was somewhat lower. Contrary to expectations based on the nearly distinct distribution of the two groups of haplotypes (Figure 1), there was slightly more variation among regions for the estimates based on haplotype frequencies only (Table 2), indicating no significant phylogeographic structure. This can, however, be explained by a large amount of homoplasy in length variation, which represented a considerable part of the total variation in both African groups. There was no differentiation among mountains within the northern and the southwestern group (group 19). In the eastern group (group A), the proportion of variation among mountains was significant; however, the GST/NST test was not. The two haplotypes from the Bale Mts (B and D) were observed almost only there, making this locality distinct (Figure 1). But differentiation was significant within this group also excluding the Bale Mts (32.1% of variation among mountains, P=0.007).
The NCPA revealed a significant association between genealogy and geography only in three clades (Figure 3). For clade 1-2, this association could not be attributed to a particular process (inconclusive outcome, Table 3). For clade 2-1, the inference key indicated restricted gene flow with isolation by distance, and for the total network, past fragmentation and/or long-distance colonization (Table 3).
Because it is likely that length variation in the poly-C and poly-T stretch mutates at a higher rate than the rest of the sequence, we excluded this segment for the estimation of the mismatch distribution. For the whole African clade, the sudden expansion model was rejected with P<0.001, which is consistent with the subdivision into two distinct phylogeographic groups. The mode of the distribution was one, as expected from the nearly equal frequencies of the two most common haplotypes A and 19, which differed by one substitution (Figure 4). We also carried out the analysis for each phylogeographic group separately, although there was very little variation in each group (Figure 4). For both distributions, the sudden expansion model was not rejected. The parameter τ=2 μt was estimated as 0.368 for group A (CI 0.024–0.530 for α=0.1). For group 19, the estimate was 3.0 (CI 0.399–3.187 for α=0.1), but this value does not correspond at all to expectations from the total amount of variation observed in this group (which was quite similar to that of the group A) and will be disregarded in the following discussion as an artefact of the low sample size and level of variation.
Discussion
Our results confirm the presence of two divergent lineages of A. alpina in Africa as reported by Koch et al. (2006). In addition to the widespread African lineage, haplotypes of the Asian lineage were observed in the northernmost and the easternmost mountains in this study (Simen Mts and Gara Muleta in Ethiopia). These are the localities closest to the Arabian Peninsula. We observed only two haplotypes belonging to the Asian lineage (haplotypes 4 and G), which differed by one substitution. The diversity of this lineage in Africa was thus considerably lower than for the African lineage (nine haplotypes). All together, the occurrence of this lineage close to the Arabian Peninsula and its low diversity support the conclusion of Koch et al. (2006) about the later arrival of this clade to Africa. Interestingly, the Simen Mts and Gara Muleta are situated on opposite sides of the Great Rift Valley, suggesting either two separate immigration events from the Arabian Peninsula or recent long-distance dispersal of the Asian lineage.
The African lineage was widespread and represented in all localities except Gara Muleta. Within this lineage we found a subdivision into at least two phylogeographic groups, group A and group 19. Although from a phylogenetic point of view there was little evidence for these groups (the clade containing A had a low statistical support, and the group containing 19 was unresolved), their topology on the haplotype network and nearly distinct geographic distribution clearly support their existence in the context of a recent phylogeographic pattern. These groups were found on opposite sides of the Great Rift Valley in Ethiopia (except for haplotype C which also occurred in the Simen Mts), and extended southwards on their respective sides. In the south, group 19 extended across the Great Rift Valley to Mt Meru and Mt Kilimanjaro on the eastern side, and one haplotype of this group also occurred further north in Mt Kenya, where it co-occurred with haplotypes of group A. Thus, despite the distinct ranges occupied by the three phylogeographic groups, secondary contacts were observed both in the south (Mt Kenya) and in the north (Simen Mts; Figure 1). These secondary contacts were each represented by one haplotype found at a locality dominated by another group. It is interesting to notice that the three haplotypes forming the secondary contacts were derived haplotypes, contrary to expectations based on the observed haplotype frequencies.
Both the NST/GST test and the NCPA showed that there was a significant phylogeographic structure within Africa. The NST/GST test detected a significant association between phylogeny and geography only between the Asian and the African clade, and not between the two phylogeographic groups within the African clade, probably because of a considerable amount of homoplasy in length variation within this clade. The NCPA indicated that the significant phylogeographic pattern among the three main clades in the nested design of the network could be explained by past fragmentation and/or long-distance colonization. For the distinction between the Asian and the African clade, past fragmentation was suggested by the long branch separating the two clades on the network. This is consistent with the interpretation of Koch et al. (2006), stating that the two clades have evolved in different regions, and co-occur nowadays in Africa as a result of secondary contact resulting from a later immigration of haplotypes of the Asian clade to northeastern Africa. According to Templeton's (2004) inference key, the low divergence between the two African clades (3-1 and 3-2 in the nested design, Figure 3) indicated colonization or perhaps recent fragmentation.
The importance of the Great Rift Valley as a phylogeographic barrier has been repeatedly documented for animals, notably the Ethiopian wolf (Gottelli et al., 2004), the ostrich (Freitag and Robinson, 1993) and the wildebeest (Arctander et al., 1999). Also, phytogeographic studies have previously documented a stronger link of the Simen Mountains to the Arabian Peninsula than to the Bale Mountains (Gillett, 1955; Puff and Nemomissa, 2001), confirming the strong barrier effect of the Rift. However, for example for the Ethiopian wolf, the barrier effect was not absolute, and some dispersal across the Rift has been inferred from genetic data (Gottelli et al., 2004). The understanding of the Great Rift Valley as a significant, but permeable, barrier to dispersal may be applicable to afro-alpine plants as well. In Ethiopia, the maximum elevation in the Rift Valley is about 1600 m. Thus, assuming that the afro-alpine vegetation belt was 1000–1500 m lower during cooler periods in the Pleistocene, it would not have reached the bottom of the valley. The possibility that high-altitude grassland reached the highest parts of the floor of the Rift Valley at some time during the Pleistocene has, however, been suggested by Yalden and Largen (1992). Still, the Rift clearly represented a discontinuity in the alpine habitat for most of the Pleistocene.
The distinct phylogeographic groups observed in the Ethiopian highlands on different sides of the Rift Valley are consistent with our expectations assuming a larger and more continuous afro-alpine habitat during cooler periods of the Pleistocene, which, however, was divided by the Rift Valley. Also in the southern mountains, haplotypes belonging to the same group were observed on adjacent mountains (Meru and Kilimanjaro versus Kenya and Aberdares), as predicted assuming gradual dispersal during periods when the afro-alpine habitat was more widespread than today.
Despite the clear overall phylogeographic structure in African A. alpina, indicating gradual expansion, isolation and rather strong dispersal barriers, our results suggest several long-distance dispersal events. Three of these crossed the Great Rift Valley: (1) the Asian lineage: the Simen Mts – Gara Muleta (or two independent immigrations from the Arabian Peninsula), (2) phylogeographic group A: from Mt Kaka to the Simen Mts, (3) phylogeographic group 19: across the Great Rift Valley in the south (our data do not allow to exclude gradual dispersal over the Rift Valley in the past for this group, a scenario that we, however, consider unlikely for ecological reasons). These observations suggest that in addition to more or less gradual or step-wise range expansion during cooler periods of the Pleistocene, long-distance dispersal events have also been important for colonization of isolated mountain tops. These events were not identified by NCPA, confirming that they were not the main factors shaping the genetic pattern we observed today. Long-distance dispersal parallel to the Rift Valley may also have been necessary for the dispersal of the phylogeographic groups between Ethiopia and the southern mountain massifs.
Koch et al. (2006) dated the divergence between the three major lineages of A. alpina to the Pleistocene (about 500 000 years ago). The divergence between the two African phylogeographic groups was about four times less than between the European and African lineage (1.02 versus 4 base pairs in net number of pair-wise differences between groups, excluding length variation), clearly indicating a more recent subdivision. The mismatch distribution was consistent with a population expansion in both phylogeographic groups (however, the power to reject such a model was low). The estimate for τ=2 μt was 0.368 for group A, indicating an expansion that is twice as recent as for the European lineage (τ=0.7, Koch et al., 2006). It could not be estimated in a meaningful way for group 19, but the level of variation and the histogram of the mismatch distribution were very similar to that of group A, indicating similar ages. For the European clade, Koch et al. (2006) suggested an expansion predating the last ice age, possibly during the previous ice age. For the African lineage, the smaller estimate of τ may indicate expansions on each side of the Rift during the last ice age. Although these estimates may suggest roughly where in history to place an event, it is important to remember that they are very imprecise and that CIs are large. Previous to the inferred expansion, during the previous warm period (interglacial), A. alpina may have been restricted to at least two refugia, one on the eastern side of the Rift Valley, possibly in Ethiopia, and one on the western side or in the Kilimanjaro-Meru area in the south. Considering the distribution of diversity within lineages, the refugium of group 19 may have been in the Kilimajaro-Meru area in the south, and the refugium of group A in Ethiopia.
Our results thus suggest the following scenario for A. alpina in eastern Africa. During an early glaciation in the Pleistocene, the first A. alpina colonized Africa from the north, possibly from the Middle East via the mountains bordering the Red Sea. A first range expansion may have been followed by several cycles of range contractions and expansions in response to the Pleistocene climate fluctuations. During the previous warm period, the African lineage may have survived in different refugia in agreement with the recent fragmentation among the two African clades indicated by NCPA. In the following cool period, corresponding to the last glaciation in the northern Hemisphere, the phylogeographic groups expanded from these refugia following the increasing area of the afro-alpine habitat. At the same time, populations belonging to the Asian clade spread southwards along the mountains bordering the Red Sea and immigrated into northern and eastern Ethiopia. Thus, our data indicate that the cooler periods of the Pleistocene may have played a crucial role for the dispersal of afro-alpine plants between isolated mountain tops. However, despite the considerable barriers imposed by lowland habitats, in particular the Rift Valley, occasional long-distance dispersal also occurred among the isolated ‘sky islands’, possibly by wind or birds.
Accession codes
References
Arctander P, Johansen C, Coutellec-Vreto MA (1999). Phylogeography of three closely related African bovids (tribe Alcelaphini). Mol Biol Evol 16: 1724–1739.
Bandelt HJ, Forster P, Röhl A (1999). Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16: 37–48.
Bonnefille R, Roeland JC, Guiot J (1990). Temperature and rainfall estimates of the past 40 000 years in equatorial Africa. Nature 346: 347–349.
Burban C, Petit RJ, Carcreff E, Jactel H (1999). Range wide variation of the maritime pine bast scale Matsucoccus feytaudi Duc. (Homoptera: Matsucoccidae) in relation to the genetic structure of its host. Mol Ecol 8: 1593–1602.
DeMenocal PB (1995). Plio-Pleistocene African climate. Science 270: 53–58.
Dobeš C, Mitchell-Olds T, Koch M (2004). Phylogeographic analysis of extensively sympatric and highly diverse chloroplast haplotypes (trnL intron-trnF IGS) in North American Arabis drummondii, A. divaricarpa, and A. holboellii (Brassicaceae). Mol Ecol 13: 349–370.
Excoffier L, Laval G, Schneider S (2005). Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinf Online 1: 47–50.
Flenley J (1979). The Equatorial Rain Forest: A Geological History. Butterworths: London.
Freitag S, Robinson TJ (1993). Phylogeographic patterns in mitochondrial DNA of the ostrich (Struhtio camelus). Auk 110: 614–622.
Gillett JB (1955). The relation between highland floras of Ethiopia and British East Africa. Webbia 11: 456–469.
Goloboff P (1999). Analyzing large data sets in reasonable times: solutions for composite optima. Cladistics 15: 415–428.
Gottelli D, Marino J, Sillero-Zubir C, Funk S (2004). The effect of the last glacial age on speciation and population genetic structure of the endangered Ethiopian wolf (Canis simensis). Mol Ecol 13: 2275–2286.
Hedberg O (1951). Vegetation belts of East African Mountains. Svensk bot Tidskr 45: 140–202.
Hedberg O (1962). Intercontinental crosses in Arabis alpina L. Caryologia 15: 252–260.
Hedberg O (1969). Evolution and speciation in a tropical high mountain flora. Bot J Linn Soc 1: 135–148.
Hedberg O (1970). Evolution of the afroalpine flora. Biotropica 2: 16–23.
Hedberg O (1986). Origins of the Afroalpine flora. In: Vuilleumier F, Monasterio M (eds). High Altitude Tropical Biogeography. Oxford University Press: New York. pp 443–468.
Jukes T, Cantor C (1969). Evolution of protein molecules. In: Munro HN (ed). Mammalian Protein Metabolism. Academic press: New York.
Kingdon J (1990). Island Africa. Collins: London.
Koch M, Haubold B, Mitchell-Olds T (2000). Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase in Arabidopsis, Arabis, and related Genera (Brassicaceae). Mol Biol Evol 17: 1483–1498.
Koch M, Haubold B, Mitchell-Olds T (2001). Molecular systematics of the Brassicaceae – evidence from coding plastidic matK and nuclear chs sequences. Am J Bot 88: 534–544.
Koch M, Kiefer C, Ehrich D, Vogel J, Brochmann C, Mummenhoff K (2006). Three times out of Asian Minor: the phylogeography of Arabis alpina L (Brassicaceae). Mol Ecol 15: 825–839.
Livingstone DA (1993). Evolution of African climate. In: Goldblatt P (ed) Biological Relationships between African and South America. Yale University Press: New Haven, Connecticut. pp 455–472.
Mai DH (1995). Tertiäre Vegetationsgeschichte Europas. Fischer: Jena, Stuttgart, New York.
Messerli B, Winiger M, Hurni H, Kienholz H (1977). Bale Mountains: Largest Pleistocene Mountain Glacier System of Ethiopia. INQA Abstracts, Birmingham.
Moreau RE (1963). Vicissitides of the African biomes in the late Pleistocene. Procs Zool Soc London 141: 395–421.
Nei M (1987). Molecular Evolutionary Genetics. Columbia University Press: New York, NY, USA.
Petit RJ, Duminil J, Fineschi S, Hampe A, Salvini D, Vendramin GG (2005). Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol Ecol 14: 689–701.
Pons O, Petit RJ (1996). Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144: 1237–1245.
Posada D, Crandall KA, Templeton AR (2000). GeoDis: a program for the cladistic nested clade analysis of the geographical distribution of genetic haplotypes. Mol Ecol 9: 487–488.
Puff C, Nemomissa S (2001). The Simen Mountains (Ethiopia): comments on plant biodiversity, endemism, phytogeographical affinities and historical aspects. Syst Geog Plants 71: 975–991.
Rogers AR, Harpending H (1992). Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9: 552–569.
Ronquist F, Huelsenbeck JP (2003). MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574.
Schneider S, Excoffier L (1999). Estimation of past demographic parameters from the distribution of pair-wise differences when the mutation rates vary among sites. Application to human mitochondrial DNA. Genetics 152: 1079–1089.
Taberlet P, Gielly L, Pautou G, Bouvet J (1991). Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol 17: 1105–1109.
Templeton AR (2004). Statistical phylogeography: methods of evaluating and minimizing inference errors. Mol Ecol 10: 779–791.
Templeton AR, Boerwinkle E, Sing CF (1987). A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping: I. Basic theory and analysis of alcohol dehydrogenase activity in Drosophila. Genetics 117: 343–351.
Van de Peer Y, De Wachter R (1994). TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Applic Biosci 10: 569–570.
Yalden DW, Largen MJ (1992). The endemic mammals of Ethiopia. Mammal Rev 22: 115–150.
Acknowledgements
We thank M Kebede, A Muasya and E Knox for company and help during the fieldwork and two anonymous referees for helpful comments. This study was funded by The Norwegian Programme for Development, Research and Higher Education (NUFU) project 54/2003 to S Nemomissa and C Brochmann (‘AFROALP – Afroalpine ‘islands’ as natural laboratories: dynamics and units of plant biodiversity') and partly by grant 150322/720 to C Brochmann from the Research Council of Norway.
Author information
Authors and Affiliations
Corresponding author
Appendix A
Appendix A
Mountain region, population number (Pop), altitude in metres, geographical coordinates in degrees, date of collection, individual number (Ind) and observed haploptye for the 66 individuals of A. alpina sequenced for the cpDNA trnL intron and trnL-trnF intergenic spacer.
Rights and permissions
About this article
Cite this article
Assefa, A., Ehrich, D., Taberlet, P. et al. Pleistocene colonization of afro-alpine ‘sky islands’ by the arctic-alpine Arabis alpina. Heredity 99, 133–142 (2007). https://doi.org/10.1038/sj.hdy.6800974
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800974
Keywords
This article is cited by
-
Afro-alpine flagships revisited II: elucidating the evolutionary relationships and species boundaries in the giant senecios (Dendrosenecio, Asteraceae)
Alpine Botany (2022)
-
History and evolution of the afroalpine flora: in the footsteps of Olov Hedberg
Alpine Botany (2022)
-
Colonization of the Southern Hemisphere by Sagina and Colobanthus (Caryophyllaceae)
Plant Systematics and Evolution (2022)
-
Diversity and endemism of the flora of Ethiopia and Eritrea: state of knowledge and future perspectives
Rendiconti Lincei. Scienze Fisiche e Naturali (2021)
-
The phylogeographic structure of Arabis alpina in the Alps shows consistent patterns across different types of molecular markers and geographic scales
Alpine Botany (2018)