Abstract
Drosophila melanogaster samples were collected from a large population in two habitats: farmyards and distilleries. Samples were taken from two villages in each habitat. Three isofemale lines were established from all four samples and full-sib crosses were set in each isofemale line. Activities of four enzymes (ADH, αGPDH, IDH and 6PGDH) were measured in the offspring of each cross on starch gel after electrophoresis. Broad sense heritabilities and additive genetic variances were estimated in all four samples.Most of the activity variation was observed within the isofemale lines. The isofemale lines tended to be more different in the distilleries than in the farmyards. There was no significant difference in the average activities between the two habitats for any of the enzymes investigated. The additive genetic variance of the enzyme activities did not exhibit a consistent habitat pattern. In the farmyard habitat, we detected a higher activity variation in Tiszafüred than in the other village. Strong correlation was observed among the activities of the enzymes investigated. Correlation coefficients indicated higher level of correlation in the samples collected in Tiszafüred than in those originating from Tiszaszőlős. The heritability values were rather high and they had a considerable variation both between the habitats and across the enzymes.
Similar content being viewed by others
Introduction
Enzyme activity is a phenotypic trait determined by genetic and environmental factors (Clark, 1989, 1990). Variation in this phenotypic trait has been extensively surveyed in Drosophila melanogaster using isogenic lines. The results show that natural D. melanogaster populations exhibit a considerable amount of genetic variation in enzyme activity (Lewis and Gibson, 1978; Laurie-Ahlberg et al, 1980, 1982). For some enzymes (eg alcohol dehydrogenase (ADH)), a large portion of this genetic variation could be attributed to regulatory genes (Laurie and Stam, 1994; Mercot, 1994). Wilton et al (1982) and Clark and Keith (1988) have detected significant correlations among the activities of enzymes involved in the same metabolic pathways, indicating their joint regulation.
Another way of studying phenotypic variation in enzyme activity in natural populations is to analyse the structure of this variation in a manner similar to that used in molecular studies (eg Snyder and Linton, 1984; Cesaroni et al, 1989; Descimon and Napolitano, 1993). In order to do so, one must be able to measure the variation among individuals (Pierce and Crawford, 1994). In our previous studies, we assessed individual variation in the activities of some enzymes in D. melanogaster by using simple gel electrophoresis and measuring the activities on the gel after the separation of the protein molecules. This method gives reproducible results (Pecsenye and Saura, 2002). At the same time, it allowed us to construct an experimental design to partition the total phenotypic variation in enzyme activities into genetic and environmental components.
Heritability values were often estimated for various traits in Drosophila species. In many cases, the additive genetic variance was determined by parent–offspring analysis (Jenkins and Hoffmann, 1994; Gibert et al, 1998; Sgro and Hoffmann, 1998). In some studies, however, heritabilities were computed by using half-sib analysis (Bubliy and Loeschcke, 2000) or the ‘intra-sire’ method (Islam and Parveen, 1992). Heritabilities of enzyme activities were also determined in D. melanogaster by using isogenic lines (Laurie-Ahlberg et al, 1982).
The main goal of the present study was to determine the additive genetic variances of the activities of some enzymes in a D. melanogaster population and to contrast these variances between samples living in two different habitats, where the alcohol concentration of the breeding substrate is markedly different. We also aimed at estimating the heritability of the activities of these enzymes. Two of the enzymes included in this study are directly associated with the metabolism of ethanol: ADH and α-glycerophosphate dehydrogenase (αGPDH) (eg Geer et al, 1985; Oudman et al, 1991; Heinstra, 1993), while the other two (isocitrate dehydrogenase (IDH) and 6-phosphogluconate dehydrogenase (6PGDH)) are involved in central energy metabolism (Dickinson and Sullivan, 1975; O'Brien and McIntyre, 1978) and, therefore, have an indirect relation to ethanol degradation. While the genetic structure of natural populations living in different habitats has been extensively surveyed, it is novel to compare the level and organisation of variation in enzyme activities among different habitats.
Methods
Samples
D. melanogaster females (P) were collected from a large population (Central Tisza region) in two habitats: farmyards and distilleries. Two villages (Tiszafüred and Tiszaszőlős) were sampled in both habitats. Several isofemale lines (IFL) were established from all four samples (two habitats × two villages). All IFLs were kept in the laboratory on cornmeal medium at 24°C.
Experimental design
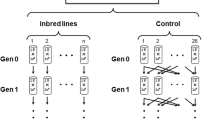
Creating the F2 generation, three full-sib crosses were set in three randomly selected isofemale lines of all four samples. For each cross, one male was put together with three females. A few days later, the male was discarded and the females were allowed to lay eggs separately. In total, 10 daughters of each female were collected and used later to determine enzyme activities (Figure 1). In this way we had 90 offsprings from each isofemale line.
Sample preparation
Offspring from the crosses
Altogether 1080 female offspring were frozen in the F2 generation. Samples of these females were prepared ‘individually’, that is, each female was homogenised separately in 50 μl of the following buffer: 0.01 M Tris/HCl (pH=7.5) and 2 mg/ml dithiotreitol. Then the homogenates were centrifuged (12 000 rpm, 10 min).
Standard strains
The Oregon-R strain kept in the Umeå Drosophila Stock Center was used as a standard strain in the study of enzyme activity variation. The standard samples were prepared ‘collectively’, ie, 70 females (approx. 7 days old) were collected from the Oregon-R strain and homogenised in 2.4 ml (30 μl/individual) of the above buffer. After homogenisation, the standard sample was centrifuged (12 000 rpm, 10 min) and the supernatant was distributed into several 60 μl aliquots. These aliquots were then frozen until electrophoresis.
Electrophoresis and enzyme staining
While the standard samples were applied at fixed positions on the gels (1, 7, 13 and 25) the samples from the crosses were randomly arranged. Nevertheless, there was some regularity in the application of these natural samples: (i) all the 10 offspring from a cross were put on the same gel; (ii) one gel contained the offspring of crosses from both habitats randomly combined (eg TS FY 1/2+TF DI 2/3). From all samples, 10 μl of the supernatant was applied on the gels.
The starch gel electrophoresis was always carried out in the same way throughout the whole study. Electrode buffer: 0.28 M Tris–86 mM citric acid (pH=7.5); gel buffer: 1:28 dilution of the electrode buffer; running conditions: 14 V/cm, 6–7 C, 3.5 h. After electrophoresis the gel was sliced and four different slices were systematically stained for the four enzymes.
The staining procedure was kept identical throughout the study. All four enzymes were stained for a specific length of time. This period was chosen to lay within the linear phase of the activity of the enzymes, as was ascertained in a set of preliminary studies. ADH assay: 0.1 M Tris-HCl (pH=8.5), 1 mM EDTA, 0.6 mM NAD, 0.25 mM nitroblue tetrazolium (NBT), 0.1 mM phenazine methosulphate (PMS), 2% ethanol, 2% butanol; 40 min at 37°C in the dark. αGPDH assay: 0.1 M Tris-HCl (pH=8.5), 1 mM EDTA, 0.6 mM NAD, 0.25 mM NBT, 0.1 mM PMS, 12 mM DL-α-glycerophosphate; 30 min at 37°C in the dark. IDH assay: 0.1 M Tris-HCl (pH=8.5), 5 mM MgCl2, 0.25 mM NADP, 0.25 mM NBT, 0.1 mM PMS, 4 mM DL-isocitric acid; 40 min at 37°C in the dark. 6PGDH assay: 0.1 M Tris-HCl (pH=8.0), 5 mM MgCl2, 0.25 mM NADP, 0.25 mM NBT, 0.1 mM PMS, 2 mM 6-phosphogluconic acid; 60 min at 37°C in the dark.
After staining, the gels were immersed in distilled water and photographed immediately. The photographic negatives were scanned with Scanmaker E6 and the images of the gels were analysed by the Gel Pro TM analyzer 2.0 programme package. For each sample, the enzyme activities were characterised by the sum of the total optical densities of the individual bands.
Statistical procedures
The data were analysed with generalised linear models under the assumption of a constant coefficient of variation, that is, we specified gamma error distribution coupled with inverse link function (see McCullagh and Nelder, 1989).
In the first part of the analyses, a standardisation procedure was carried out. Enzyme activities measured in the standard samples (standard activities) were used in this process. The first models constructed for the standard data only contained gel as main factor (Crawley, 1993; Francis et al, 1994). Based on these models we calculated special weights (gel weights) for the different enzymes to account for the variation among the gels. The next step was to test whether the position of the samples within the gels had any effect on the activities. Except for 6PGDH, there was a clear correlation between the gel weighted standard activity values and the position of the samples on the gels. Three different regression models were constructed for the three enzymes with sample positions as independent variables and the gel-weighted standard activity values as dependent variables. The slopes of the different regression lines were then used to calculate the position weights for all three enzymes to account for the activity differences due to the positions of the samples on the gels (for further details see Pecsenye, 1998).
In the second part of the analyses, the original activity values of all enzymes were corrected for the differences among the gels using the gel weights. Then the gel weighted activity data for αGPDH, ADH and IDH were further corrected using the position weights. These standardised activity data were then used in a series of nested analyses of deviance. The highest level of the hierarchy was the sample, isofemale lines were nested within samples, males were nested within isofemale lines and females were nested within males. Separate analyses were carried out for the samples collected in the two habitats. The analyses were performed by the GLIM4 programme package.
In order to analyse the interaction among the enzymes investigated product-moment correlation coefficients were estimated using generalised linear models (Crawley, 1993). The data set contained the activity values of each individual for all four enzymes and the computation was carried out separately for the samples originating from the four locations (two habitats × two villages).
In the third part of study, the traits were analysed for each sample with the following model: trait=line, sire nested within line, and dam nested within line and sire interaction using the mixed model least-squares and maximum likelihood program of Harvey. As progenies of different F1 dams were treated in the same environment (temperature, diet) no common environmental effect was assumed within the family. The additive genetic variance expected in the between-family variance component was 0.25, the between-dams expected component was 0.25, and the expected within-family variance component was 0.5. Variance components were estimated by the same model using Henderson's method 3 (Henderson, 1953; Harvey, 1990). Estimates of broad sense heritabilities were calculated using the following formula:

The standard errors of the heritability estimates were calculated.
Results
The average activities of all four enzymes had high variation in all samples (Figure 2). No significant activity differences were detected between the two habitats in any of the two villages/samples for any of the four enzymes (Figure 2). The distribution of variation did not exhibit a clear habitat specific pattern. The highest portion of deviance was explained by the differences among the offspring within a cross for all enzymes in both habitats (Table 1: WFm). The level of differentiation tended to be slightly higher among the distillery samples than among the farmyard ones. For ADH and IDH, the differences between the samples accounted for a clearly higher portion of the total deviance in the distillery habitat compared to the farmyards (Table 1, BS: ADH – 2.7 vs 0.6; IDH – 2.4 vs 0.1). For αGPDH and 6PGDH, however, it was the portion of variation attributable to the differences between the IFLs that was evidently higher in the distillery samples than in the farmyard ones (Table 1, WS=BIFL: for αGPDH – 8.5 vs 1.0; for 6PGDH – 4.5 vs 0.1).
Additive genetic variances did not exhibit a consistent habitat pattern across the four enzymes. For IDH and 6PGDH, the differences in the additive component of genetic variation between the two habitats were similar in the two sample sites (Table 2: VA). Nevertheless for IDH, this variance was higher in the distillery samples compared to the farmyard ones, while for 6PGDH it was higher in the farmyard samples than in the distillery ones. The most striking difference between the two samples sites was observed for ADH. In Tiszafüred, the variance in ADH activity was significantly higher in the distillery samples than in the farmyard ones, while it was exactly the other way around in Tiszaszőlős (Table 2: VA).
It is remarkable that the farmyard samples originating from Tiszafüred tended to have higher additive genetic variance in enzyme activity than those of Tiszaszőlős except for ADH. In our earlier study (Pecsenye, unpublished), we analysed the level and structure of enzyme polymorphism in several samples collected in these two villages. Both the parameters of polymorphism and the results of F-statistics suggested that the level genetic variation was higher in Tiszafüred than in Tiszaszőlős (Table 3).
The broad sense heritability values of enzyme activities estimated for the four samples were all significantly different from zero. These heritability values varied between 0.37 and 0.87 for the different enzymes in the four samples without any consistent habitat pattern across the two villages (Table 2: H2).
The results of the correlation studies indicated that in general, the enzymes investigated were strongly correlated with each other (Table 4). The pattern of correlations, however, was slightly different in the four samples. Nonetheless, ADH and IDH were always strongly correlated, while the association between αGPDH and 6PGDH was usually very weak. Enzyme activities seemed to be more strongly correlated in the samples collected in Tiszafüred than in those originating from Tiszaszőlős. In Tiszafüred, the association among the activities of the enzymes investigated appeared to be stronger in the farmyard samples than in the distillery ones.
Discussion
Farmyards and distilleries are markedly different habitats. While farmyards are natural or seminatural habitats of the fruit flies, distilleries can be considered as artificial. Farmyard is the permanent habitat of the flies, while distillery communities are founded by hundreds of larvae and pupae taken with the barrels of mash from the farmyards to the distillery. Farmyards have, in general, low ethanol concentration in the breeding substrates (up to 3–4%). In distilleries, however, the ethanol concentration can be extremely high in the fermenting mash (close to 15%). All these features suggest two possible evolutionary forces affecting the communities living in distilleries. On the one hand, selection must be stronger in distilleries than in the surrounding farmyards due to the high ethanol concentration in the breeding substrate. But on the other hand, genetic drift can also be very efficient in distilleries as a consequence of repeated founder effects and great fluctuations in population size.
In some of our earlier studies, we were searching for adaptive differences between farmyards and distilleries. In Pecsenye and Meglècz (1995), we compared the genetic structure of samples collected in these two habitats. We could not detect any difference between samples collected in distilleries and farmyards in their allele frequency distribution at any of the loci investigated. Thus our results supported those surveys, where no significant differences were found in AdhF allele frequency among habitats with different alcohol concentration in the breeding substrate (eg Oakeshott et al, 1984). In two further surveys (Pecsenye et al, 2002; K Pecsenye and A Saura, unpublished), we found that ADH activities and alcohol tolerance were similar in distillery and farmyard samples both in the adult and in the larval stages. In other words, the distillery samples, which originated from a habitat with fairly high ethanol concentration, had as high an ADH activity and were as sensitive to ethanol as the farmyard samples. This finding is especially interesting in the light of the literature. Although in natural populations, ADH activity did not always correlate with the alcohol content of the breeding substrate (Barbancho et al, 1987; Mercot and Massaad, 1989) alcohol tolerance was always higher in habitats with high alcohol concentration, for example, in wineries (eg Hickey and McLean, 1980; Gibson and Wilks, 1988). The outcome of the present study is in agreement with our earlier results. That is, no adaptive differences were observed either in ADH or in αGPDH activities between the two habitats. At the same time, we detected a slightly higher level of differentiation between the distillery samples compared to the farmyard ones. Accordingly, we again concluded that genetic drift was a much more efficient evolutionary force than selection in distilleries.
Considering the farmyard habitat only, the additive genetic variance was higher for most of the traits investigated in Tiszafüred than in Tiszaszőlős. This finding is in agreement with our earlier results on the level of genetic variation in these sample sites (Pecsenye and Meglècz, 1995). It suggests that the distribution of variation might be uneven within a large population. There might exist local demes where variation is accumulating, while in others it is declining. Our results suggest that the local deme in Tiszafüred exhibits a relatively higher level of variation than the one in Tiszaszőlős. At the same time, the association among enzyme activities was stronger in Tiszafüred than in Tiszaszőlős. It thus appears that the higher level of activity variation generates a more efficient correlation among the enzymes.
Since founder individuals from the farmyards established distillery communities practically independently in the two villages, this process can be regarded as two parallel sampling procedures. Considering this scenario, it is evident that there cannot be two similar samplings in terms of the consequences of the process. In other words, assuming founder effects we expect different deviations between the two habitats in the two villages. In Tiszafüred, the additive genetic variance for ADH increased in the distillery samples compared to the farmyard ones. In contrast, genetic variance of this enzyme was lower in the distillery habitat than in the farmyard in Tiszaszőlős. The correlation between ADH and αGPDH was maintained in the distillery habitat in Tiszaszőlős, while it diminished in Tiszafüred. These results seem to support the assumption of independent founder events in the two villages.
Several authors have reported on the heritabilities of various morphological and fitness related traits in different Drosophila species (eg Jenkins and Hoffmann, 1994; Imasheva et al, 1999; Bubliy and Loeschcke, 2000). The general conclusion of these studies is that morphological traits express higher heritabilities than fitness traits (Islam and Parveen, 1992; Gibert et al, 1998). One possible reason behind this phenomenon could be that fitness related traits ought to have the highest level of phenotypic plasticity to facilitate adaptation in a changing environment (Willis, 1991). A high heritability value, however, suggests that the given trait is mostly determined by genetic factors and, therefore, it has a low level of phenotypic plasticity. Nevertheless, larger phenotypic plasticity in itself does not imply more efficient adaptation, the amount of genetic variation also affects adaptivity (Donovan and Ehleringer, 1994; Wolff et al, 2001).
Heritabilities and additive genetic variances have rarely been estimated for enzyme activities. In Laurie-Ahlberg et al (1982), the broad sense heritabilities were estimated for 24 enzymes in second and third chromosome isogenic lines. The four enzymes we investigated in this study were also involved in that by Laurie-Ahlberg et al (1982). The heritability values estimated by Laurie-Ahlberg et al (1982) varied around 0.4–0.9 and they differed greatly in the second and third chromosome isogenic lines. Our results are in agreement with theirs; heritability values fluctuated around 0.4––0.8 and exhibited great variation among the samples. It is interesting that the heritability values of enzyme activities seem to be intermediate between those of morphological and fitness-related traits. Thus, the phenotypic plasticity of enzyme activities can also be expected to be fairly high. It is not surprising if we consider that most enzymes are associated with each other in the intermediate metabolism, which in turn results in a considerable level of phenotypic plasticity. The strong overall correlation among enzyme activities was also detected in this study.
References
Barbancho M, Sanchez-Canete FJS, Dorado G, Pinesda M (1987). Relation between tolerance to ethanol and alcohol dehydrogenase (ADH) activity in Drosophila melanogaster: selection, genotype and sex effects. Heredity 58: 443–450.
Bubliy OA, Loeschcke V (2000). High stressful temperature and genetic variation of five quantitative traits in Drosophila melanogaster. Genetica 110: 79–85.
Clark AG (1989). Causes and consequences of variation in energy storage in Drosophila melanogaster. Genetics 123: 131–144.
Clark AG (1990). Genetic components of variation in energy storage in Drosophila melanogaster. Evolution 44: 637–650.
Clark AG, Keith LE (1988). Variation among extracted lines of Drosophila melanogaster in triacylglycerol and carbohydrate storage. Genetics 119: 595–607.
Cesaroni D, Allegrucci G, Angelici MC, Racheli T, Sbordoni V (1989). Allozymic and morphometric analysis of populations in the Zygaena purpuralis complex (Lepidoptera, Zygaenidae). Biol J Linn Soc 36: 271–280.
Crawley MJ (1993). GLIM for Ecologists. Blackwell Scientific Publications: London.
Descimon H, Napolitano M (1993). Enzyme polymorphism, wing pattern variability and geographical isolation in an endangered butterfly species. Biol Conserv 66: 117–123.
Dickinson WJ, Sullivan DT (1975). Gene–Enzyme Systems in Drosophila. Springer-Verlag: Berlin.
Donovan A, Ehleringer JR (1994). Potential for selection on plants for water-use efficiency as estimated by carbon isotope discrimination. Am J Bot 81: 927–935.
Francis B, Green M, Payne C (1994). GLIM 4. The Statistical System for Generalised Linear Interactive Modelling. University Press Inc.: New York.
Geer BW, Langevin ML, McKechnie SW (1985). Dietary ethanol and lipid synthesis in Drosophila melanogaster. Biochem Genet 23: 607–622.
Gibert P, Moreteau B, Moreteau JC, David JR (1998). Genetic variability of quantitative traits in Drosophila melanogaster (fruit fly) natural populations: analysis of wild-living flies and of several laboratory generations. Heredity 80: 326–335.
Gibson JB, Wilks AV (1988). The alcohol dehydrogenase polymorphism of Drosophila melanogaster in relation to environmental ethanol, ethanol tolerance and alcohol dehydrogenase activity. Heredity 60: 403–414.
Harvey W (1990). Users's Guide for LSMLMW and MIXMDL with PARMCARD, in press.
Heinstra PWH (1993). Evolutionary genetics of the Drosophila alcohol dehydrogenase gene–nzyme system. Genetica 92: 1–22.
Henderson CR (1953). Estimation of variance and covariance components. Biometrics 9: 226–252.
Hickey DA, McLean MD (1980). Selection for ethanol tolerance and Adh allozymes in natural populations of Drosophila melanogaster. Genet Res Cambridge 36: 11–15.
Imasheva AG, Bosenko DV, Bubli OA (1999). Variation in morphological traits of Drosophila melanogaster (fruit fly) under nutritional stress. Heredity 82: 187–192.
Islam MS, Parveen S (1992). Estimates of heritability for quantitative traits in Drosophila melanogaster. Pak J Zool 24: 105–109.
Jenkins NL, Hoffmann AA (1994). Genetic and maternal variation for heat resistance in Drosophila from the field. Genetics 137: 783–789.
Laurie CC, Stam LF (1994). The effect of an intronic polymorphism on alcohol dehydrogenase expression in Drosophila melanogaster. Genetics 138: 379–385.
Laurie-Ahlberg CC, Wilton AN, Courtsinger JW, Emigh TH (1982). Naturally occuring enzyme activity variation in Drosophila melanogaster. I. Sources of variation for 23 enzymes. Genetics 102: 191–220.
Laurie-Ahlberg CC, Maroni G, Bewley GC, Lucchesi JC, Weir BS (1980). Quantitative genetic variation of enzyme activities in natural populations of Drosophila melanogaster. Proc Natl Acad Sci USA 77: 1073–1077.
Lewis N, Gibson J (1978). Variation in amount of enzyme protein in natural populations. Biochem Genet 16: 159–170.
McCullagh P, Nelder JA (1989). Generalized Linear Models 2nd edn. Chapman & Hall: London.
Mercot H (1994). Phenotypic expression of ADH regulatory genes in Drosophila melanogaster: a comparative study between a palearctic and a tropical population. Genetica 94: 37–41.
Mercot H, Massaad L (1989). ADH activity and ethanol tolerance in third chromosome substitution lines in Drosophila melanogaster. Heredity 62: 35–44.
Oakeshott JG, Gibson JB, Wilson SR (1984). Selective effects of the genetic background and ethanol on the alcohol dehydrogenase polymorphism in Drosophila melanogaster. Heredity 53: 51–67.
O'Brien SJ, MacIntyre RJ (1978). Genetics and biochemistry of enzymes and specific proteins of Drosophila. In: Ashburner M, Wright TRF (eds) The Genetics and Biology of Drosophila. Academic Press: London, Vol, 2a pp 395–551.
Oudman L, van Delden W, Kamping A, Bijlsma R (1991). Polymorphism at the Adh and Gpdh loci in Drosophila melanogaster: effects of rearing temperature on developmental rate, body weight, and some biochemical parameters. Heredity 67: 103–115.
Pecsenye K (1998). Detection of individual variation in enzyme activity in natural populations of Drosophila melanogaster. Hereditas 128: 145–151.
Pecsenye K, Meglécz E (1995). Enzyme polymorphism in Drosophila melanogaster populations collected in two different habitats in Hungary. Genetica 96: 257–268.
Pecsenye K, Saura A (2002). Structure of variation in enzyme activity in natural Drosophila melanogaster populations. Hereditas 136: 75–83.
Pecsenye K, Mester V, Bokor K (2002). Distillery as a special habitat for Drosophila melanogaster. Acta Biologica Debrecina 24: 160–176.
Pierce VA, Crawford DL (1994). Rapid enzyme assays investigating the variation in the glycolytic pathway in field-caught populations of Fundulus heteroclitus. Biochem Genet 32: 315–330.
Sgro CM, Hoffmann AA (1998). Heritable variation for fecundity in field-collected Drosophila melanogaster and their offspring reared under different environmental temperatures. Evolution 52: 134–143.
Snyder TP, Linton MC (1984). Population structure in black flies: allozymic and morphological estimates for Prosimulium fuscum and P. mixtum (Diptera: Simuliidae). Evolution 8: 942–956.
Willis MB (1991). Dalton's Introduction to Practical Animal Breeding. 3rd edn. Blackwell Scientific Publications: Oxford.
Wilton AN, Laurie-Ahlberg CC, Emigh TH, Courtsinger JW (1982). Naturally occuring enzyme activity variation in Drosophila melanogaster. II. Relationships among enzymes. Genetics 102: 207–221.
Wolff R, Abbott L, Pistorale S (2001). Reproductive strategy of Bromus catharticus Vahl (cebadilla criolla): phenotypic plasticity in natural population progenies. J Genet Breed 55: 67–74.
Acknowledgements
We are very grateful to Professor Zoltán Varga for his helpful comments throughout this study. The technical assistance of Valéria Mester is highly appreciated. The project was supported by OTKA 16336 and 30528 research grants of the Hungarian Academy of Sciences.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pecsenye, K., Komlósi, I. & Saura, A. Heritabilities and additive genetic variances of the activities of some enzymes in Drosophila melanogaster populations living in different habitats. Heredity 93, 215–221 (2004). https://doi.org/10.1038/sj.hdy.6800497
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800497
Keywords
This article is cited by
-
Functional significance of genetic polymorphisms
Frontiers of Biology in China (2009)
-
Genetics Analysis of Larval Foraging Behavior in Drosophila Funebris
Behavior Genetics (2008)