Abstract
Anaecypris hispanica is a cyprinid fish which is endemic to the Guadiana River basin in the Iberian Peninsula, and whose abundance and geographical range have contracted considerably during the last 20 years. We investigated mitochondrial DNA cytochrome b and control region variation among specimens representative of nine tributaries, using direct sequencing and diagnostic restriction fragment length polymorphism. The samples from the Caia, Degebe, Ardila, and Odeleite rivers exhibited haplotypes that differed by a large number of site differences, which may be indicative of population bottlenecks that have caused stochastic extinction of haplotypes. In contrast, the populations from the Xévora, Chança, Carreiras, Vascão and Foupana rivers exhibited low levels of nucleotide diversity, which together with high haplotype diversity may also be indicative of genetic bottleneck events, with subsequent population expansion. Phylogenetic analyses, a minimum spanning network, and an analysis of molecular variance revealed geographical structuring, suggesting limited or no gene flow between populations. The populations from extreme southern rivers (Foupana and Odeleite) are monophyletic entities, suggesting that they have been isolated, probably as a consequence of brackish water upstream of their confluence with the Guadiana. The results suggest that the Foupana and the Odeleite populations, and the remaining northern populations altogether should be managed as three distinct Evolutionary Significant Units (ESUs). Within the northern ESU, four Management Units (MUs) should be considered.
Similar content being viewed by others
Introduction
The Cyprinidae is the most abundant freshwater fish family in the Iberian Peninsula, containing more species and endemic taxa than any other family. The high level of endemics, also observed in other groups of terrestrial vertebrates, is probably due to the geographical isolation of the Iberian Peninsula by the Pyrenees and to its climatic conditions (Almaça, 1976).
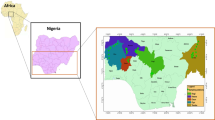
Many elements of this fish fauna have declined over the last two decades, mainly as a consequence of habitat degradation by impoundment and river regulation, sand extraction, pollution and introduction of exotic fishes (Almaça, 1995; Elvira, 1995; Collares-Pereira et al., 2000a, b; Cowx & Collares-Pereira, 2000). Anaecypris hispanica (Steindachner 1866) is presently the most threatened Iberian primary freshwater fish (SNPRCN, 1991; Blanco & González, 1992). This cyprinid is restricted to the Guadiana River drainage (Fig. 1 ), where it was abundant in the Portuguese section. However, its abundance and geographical range have contracted dramatically during the last 20 years (Collares-Pereira et al., 1998, 1999; I. Doadrio and B. Elvira, unpubl.), and Portuguese populations were considered to be fragmented into seven nuclei (Fig. 1) by Collares-Pereira et al. (1999). Recent data (Collares-Pereira et al., 2000b) suggest that the species is more abundant in the Caia, Chança, Vascão, and Odeleite rivers, and less common in the Xévora and Carreiras rivers. Within rivers, A. hispanica is patchily distributed, preferring small, shallow, well oxygenated streams, with aquatic and riparian vegetation, coarse substrate and medium to low flow. The fish seems to migrate upstream to spawn, with spawning activities restricted to spring and early summer (Ribeiro et al., 2000).
(A) Distribution area of Anaecypris hispanica in the Iberian Peninsula; (B) The Guadiana River basin in Portugal. N1–N7 represent groupings of sites (nuclei) where A. hispanica was found in 1997 (Collares-Pereira et al., 1999), and (•) indicates collecting sites.
The Guadiana River drainage exhibits a typical Mediterranean hydrological regime, characterized by extensive seasonal and annual fluctuations. Floods often occur in the wet season, while in the long dry season streams often have sections that lack continuous surface water, being composed of a series of isolated pools. The Portuguese tributaries of Guadiana within the distribution area of A. hispanica exhibit mean annual flows that range from 20 × 103 m3 in Xévora to 516 × 103 m3 in Ardila, and periods of desiccation that vary from 0.3 month in Vascão to 4.6 months in Xévora (INAG, 1996, 1997).
The progressive decline and fragmentation of A. hispanica populations identifies the need for the development of a recovery plan. The relevance of genetic information to species conservation planning has long been recognized (e.g. Lande & Barrowclough, 1987; Simberloff, 1988), and population genetic information has assumed an important role in conservation biology. Estimates of genetic variation within and between populations can provide important information on the level of interaction between local populations and permit assessment of the contribution of a metapopulation structure to regional persistence (reviewed in Hanski, 1999). Molecular markers are also an important tool for identifying population units that merit separate management and high priority for conservation. The definition of independent units for conservation of most widespread use in the last few years is the one of Moritz (1994), although recently it has become a point of debate (Paetkau, 1999; Crandall et al., 2000; Goldstein et al., 2000). Moritz distinguished two types of conservation units, namely management units (MUs), representing populations that are demographically independent, and evolutionary significant units (ESUs), which represent historically isolated sets of populations that are on independent evolutionary trajectories. ESUs are recognized by reciprocal monophyly for mitochondrial DNA (mtDNA) alleles, whereas MUs are recognized by significant divergence in allele frequencies.
The aim of the present study was to use mtDNA variation in A. hispanica to identify genetic units for conservation and to investigate the effect of population reduction and fragmentation on the distribution of genetic variation. We used direct sequencing and restriction fragment length polymorphism (RFLPs) of both the cytochrome (cyt) b gene and control region of specimens of A. hispanica collected from throughout its geographical range in Portugal. Results will facilitate the development of a rational programme for the conservation of this highly endangered fish.
Materials and methods
Sampling
One hundred and thirty-three specimens of A. hispanica were collected by electrofishing in nine Portuguese tributaries of the Guadiana river between 1997 and 1999 (Fig. 1 and Table 1). A portion of pelvic fin was removed and fixed in absolute ethanol. Fish were returned alive to the river.
DNA extraction and PCR amplification
Total genomic DNA was extracted following standard protocols of digestion with SDS and proteinase K, followed by phenol/chloroform extraction (Hillis et al., 1996). The cyt b gene and a segment of the control region located between the tRNApro gene and the conserved central domain were amplified by polymerase chain reaction (PCR) for each individual, using the set of primers and the conditions described by Brito et al. (1997), and Gilles et al. (2001), respectively.
Sequencing and sequence data analysis
PCR products for up to five specimens from each river (Table 1) were sequenced with an automated sequencer (Genome Express, SA). DNA sequences were aligned manually using MacDNASIS (version 2.0), and the identity of the sequenced fragments was confirmed by their alignment to cyt b and control region sequences of Cyprinus carpio (Chang et al., 1994). Sequences of both fragments were used to define ‘composite’ haplotypes.
Haplotype (h) and nucleotide diversity (π) (Nei, 1987) were calculated from the sequences, using ARLEQUIN version 2.000 (Schneider et al., 2000). Estimates of sequence divergence between haplotypes were determined with the Kimura two-parameter model (Kimura, 1980). Relationships among haplotypes were visualized using both neighbour-joining (NJ) and maximum parsimony methods as implemented in PAUP* (Swofford, 2000), using Leuciscus carolitertii and Chondrostoma willkommii as outgroups. Most parsimonious trees were obtained by heuristic search (MULPARS, TBR, 50 replicates). Support for nodes was assessed by bootstrap resampling using 1000 replicates. In addition, the number of substitutions between haplotypes was used to construct a Minimum Spanning Network (MSN, Excoffier & Smouse, 1994) as implemented in ARLEQUIN. A hierarchical analysis of population subdivision was performed using the analysis of molecular variance (AMOVA, Excoffier et al., 1992) implemented in arlequin, incorporating estimates of sequence divergence among haplotypes. The significance of the variance components and associated Φ-statistics were tested using 1000 nonparametric random permutations.
RFLP analysis
The software MacClade version 3.0 (Maddison & Maddison, 1992) was used to identify polymorphic characters (base changes) that define mtDNA lineages identified by the phylogenetic analysis. Sequences were then searched with MacDNASIS version 2.0 to identify restriction enzymes that cleaved at polymorphic sites.
PCR products of both fragments were generated for 15–21 additional specimens from each the Caia, Ardila, Chança, Vascão, and Foupana rivers (Table 1) and examined using the diagnostic enzymes (AvaII, BanI, and BfaI). Genetic variation of cyt b sequences from these specimens was further analysed using 13 additional restriction enzymes: AluI, BstNI, BstUI, DdeI, HaeIII, HhaI, HinfI, HpaII, MboI, NciI, RsaI, StyI, and TaqI. Amplified DNA (150–200 ng) was digested using 2–3 units of enzyme and following the conditions recommended by the suppliers (Amersham, GibcoBRL, New England Biolabs, and Pharmacia Biotech). Restriction fragments were separated through 2% agarose/TBE gels, stained with ethidium bromide. Fragment lengths were determined by comparison with a 100-bp molecular weight standard (Pharmacia Biotech) and each fragment profile was analysed against sequences of A. hispanica to determine the position of each restriction site change.
A hierarchical analysis of geographical partitioning of genetic variation (AMOVA) within the RFLP data set was performed, using evolutionary divergence d (Nei & Tajima, 1983).
Results
Sequence data analysis
Sequences of the entire cyt b gene and a segment of the control region (1140 and 678 bp, respectively) from 42 specimens representing nine tributaries of the Guadiana River drainage revealed 35 ‘composite’ haplotypes (Appendix), leading to a high estimate of diversity (h ± SE=0.99 ± 0.01). No haplotypes were shared by populations, with the exception of H5, which was found in one specimen from the Caia River and one specimen from the Xévora River (Fig. 2). Estimates of within-river haplotype diversity were also high (Table 2), in particular for the populations from the Caia, Xévora, Degebe, Ardila, and Vascão rivers, where each of the five individuals examined exhibited an unique haplotype. The Chança and Carreiras rivers showed the lowest estimates of haplotype diversity, with two haplotypes in five and three specimens, respectively.
Neighbour-joining tree for all haplotypes of Anaecypris hispanica, using Kimura’s two-parameter distance (Kimura, 1980). The strict consensus of the most parsimonious trees showed essentially the same topology. Numbers indicate the percentage of bootstrap replicates that support each branch node (left/right numbers refer to neighbour-joining/maximum parsimony analyses, respectively). Haplotype frequencies within the nine rivers sampled are shown (C, Caia; X, Xévora; D, Degebe; A, Ardila; CH, Chança; CR, Carreiras; V, Vascão; F, Foupana; O, Odeleite).
Haplotypes differed by 1–29 substitutions, leading to estimates of pairwise sequence divergence that ranged from 0.05% to 1.54% (average = 0.81%). The largest differences were found between the specimens from the rivers Foupana and Odeleite and all others, and the lowest was observed between individuals from the same tributary. Most haplotypes within rivers were similar (1–6 substitutions), with an average within-river pairwise divergence of 0.34%. However, some samples contained individuals with highly divergent haplotypes, namely H4 in the Caia River, H9 and H11 in the Degebe River, H14 and H17 in the Ardila River, and H32 in the Odeleite river. Accordingly, these rivers showed the highest levels of nucleotide diversity (Table 2).
Neighbour-joining (NJ) analysis of genetic divergence among haplotypes revealed four major groups (Fig. 2): group A containing all the haplotypes from the Caia, Xévora, and Chança rivers; group B comprising all the haplotypes from Degebe River and some haplotypes from the Ardila River; group C grouping all the haplotypes from the Odeleite and Foupana rivers; and group D including the remaining haplotypes found in the Ardila River, and all the haplotypes from the Carreiras and Vascão rivers. Bootstrap values were high (87–98%) for all lineages except A, which showed a bootstrap value of 52%. Exclusion of outgroups decreased the bootstrap support of this group (<50%). Within group C, two other clusters were strongly supported, one including all haplotypes from the Foupana River and the other grouping haplotypes from the Odeleite River (97 and 92%, respectively). In the NJ tree, groups B and C clustered together, and the B-C lineage was joined with group A; however, bootstrap analysis revealed that this branching pattern was not robust, occurring in less than 50% of the bootstrap replicates.
The strict consensus of the 1060 most parsimonious trees showed essentially the same topology of the NJ tree (not shown, available from M. M. Coelho). The bootstrap analysis (Fig. 2) supported the same groups as the previous analysis, with the exception of cluster A, which exhibited bootstrap values of less than 50%. The exclusion of outgroups did not have any impact on the topology of the tree.
The Minimum Spanning Network (MSN, Fig. 3) revealed that haplotypes from the same river tend to occupy the same part of the network, with the exception of the haplotypes from the Ardila river. This approach identified five groups separated by a considerable number of steps (10–13 steps), which are consistent with the groups defined in the previous analyses. Within groups, there is no clear geographical structuring.
Analysis of molecular variance (AMOVA) within the sequence data set suggested high subdivision among populations (ΦST = 0.664, P < 0.001), in keeping with the observation that all rivers exhibited unique haplotypes. Variation within rivers was also appreciable, accounting for 33.4% of the total genetic variance. In the hierarchical analysis, two geographical groups were considered, as suggested by the phylogenetic analysis among haplotypes, one group containing Caia + Xévora + Degebe + Ardila + Chança + Carreiras + Vascão and the other comprising the extreme southern populations Foupana and Odeleite. Most variance (37.4%, P=0.028) was found among groups, but a similar amount was distributed among populations within groups (36.5%, P < 0.0001), suggesting considerable genetic subdivision within each geographical area. In fact, pairwise values of ΦST among samples were in general high and significant (Table 3), the only exceptions being the estimates between the populations from the Caia and Xévora rivers, the Degebe and Ardila rivers, and the Carreiras and Vascão rivers.
RFLP analysis
The AvaII, BanI, and BfaI restriction sites diagnosed the different mtDNA lineages identified by the phylogenetic analysis: the AvaII restriction site at position 272 of the control region was found only in group B; BanI exhibited no restriction site at position 429 of cyt b in group D; and the BfaI restriction site at position 366 of cyt b was observed only in group C.
The analysis of 15–21 additional specimens from each population from the Caia, Ardila, Chança, Vascão, and Foupana rivers with the diagnostic restriction enzymes (Table 4) revealed results consistent with the phylogenetic analysis. Representatives of each sample were restricted to a single group, with the exception of Ardila, which had 12 individuals in group B, six in D, and three in A.
The analysis with 13 additional restriction enzymes revealed a total of 15 haplotypes (Table 4), the average within-river haplotype diversity being 0.548 ± 0.009. Populations from the Caia, Ardila, Chança, and Vascão rivers showed haplotype diversity estimates of the same magnitude, whereas the population from the Foupana River exhibited a considerably lower value.
The analysis of molecular variance within the RFLP data set revealed high ΦST (0.587, P < 0.0001), suggesting high subdivision among populations. Pairwise values of ΦST among samples were high and significant, with exception of the estimate between the Caia and Chança rivers (Table 3).
Discussion
Sequence data revealed, in general, high within-river haplotype diversity for A. hispanica. The only exceptions were the populations from the Chança and Carreiras rivers, which exhibited low haplotype diversity estimates. However, these low values may be due to the small number of specimens sequenced, as the additional analysis of 20 individuals from the Chança River with 16 restriction enzymes revealed that this population exhibited haplotype diversity of the same magnitude of the remaining populations.
Nucleotide diversity varied among populations. The samples from the Caia, Degebe, Ardila, and Odeleite rivers exhibited haplotypes that differed by a large number of site differences, which may be indicative of population bottlenecks that have caused stochastic extinction of some haplotypes. In contrast, the populations from the Xévora, Chança, Carreiras, Vascão and Foupana rivers exhibited low levels of nucleotide diversity. Low nucleotide diversity but high haplotype diversity may also be indicative of genetic bottleneck events, where most haplotypes became extinct, followed by population expansion. This pattern of mtDNA variation has been observed in another Iberian cyprinid species, Chondrostoma lusitanicum, that inhabits other southern Iberian catchments with a Mediterranean-type hydrological regime (Mesquita et al., in press). Analyses of molecular variance (AMOVA) with sequence and RFLP data indicate that most variation in A. hispanica is partitioned among populations, suggesting limited gene flow among populations. Even rivers with close connections (e.g. Vascão and Chança, and Foupana and Odeleite) exhibit high pairwise ΦST estimates. Only Caia and Xévora, Degebe and Ardila, and Carreiras and Vascão, which are geographically close, showed low and not significant (P > 0.05) pairwise ΦST values estimated from the sequence data, while Caia and Chança exhibited low and not significant pairwise ΦST values obtained from the RFLP data. The AMOVA algorithm occasionally returned small negative values of ΦST (e.g. the Caia–Xévora comparison), indicating that the true value is positive but small (Weir, 1996). Therefore, A. hispanica seems to possess low to moderate dispersal ability. This may be a consequence of high habitat specificity, which promotes fragmentation of populations. Little gene flow among rivers within drainages has also been observed for the cyprinid fish Tiaroga cobitis and Meda fulgida, which are restricted to certain habitats (Tibbets & Dowling, 1996).
Phylogenetic relationships among haplotypes revealed pronounced phylogenetic gaps between some branches in the gene tree, each comprising general haplotypes from geographically close rivers. Some lineages were sympatric, with the Ardila population exhibiting phylogenetically distinct mtDNA lineages. The sequence data revealed the presence of two lineages (B and D) in this population, while the RFLP data suggested an additional lineage (A). The identification of an additional lineage by RFLPs may, however, be a consequence of homoplasy of the restriction sites. The pattern of mtDNA variation in A. hispanica may be included in category II (Avise, 2000), which has been rarely found in freshwater fishes. Codistribution of phylogenetically distinct mtDNA lineages many times results from secondary admixture between allopatrically evolved populations (e.g. Dodson et al., 1995; Hurwood & Hughes, 1998). Alternatively, it may reflect the conservation of ancestral polymorphism, due to a constant large population size. Ardila is the largest tributary of the Guadiana river in Portugal and its topology allows the maintenance of deep pools, which retain water for longer; therefore, bottlenecks may have been less severe.
The population from the southern tributary Chança showed a close phylogenetic affinity with the northern Caia and Xévora populations. This pattern of mtDNA variation was unexpected as no past connections are known, and may be explained by random sorting of ancestral polymorphism (Neigel & Avise, 1986).
The Odeleite and Foupana populations are monophyletic entities. These populations may have been isolated as a consequence of brackish water upstream of the confluence of the Odeleite and Foupana tributaries with the main Guadiana River. Presently, brackish water overpasses the confluence for flows smaller than 100 m3 s–1 (Hidroprojecto/COBA/HP, 1998). Once in isolation, populations may have achieved reciprocal monophyly quickly, as bottlenecks can have a major impact on rates of divergence (Avise et al., 1984).
Importance for conservation
The genetic data suggest the presence of at least three evolutionarily significant units, ESUs (sensu Moritz, 1994): each population from the Foupana and Odeleite rivers and the remaining populations. These groups have been isolated and represent evolutionarily independent lineages. Gene flow within the northern group is also restricted. AMOVA analysis with both sequence and RFLP data indicated that the Caia and Xévora, Degebe and Ardila, and Carreiras and Vascão rivers, should be considered as discrete units. Although Caia and Chança showed a low and not significant pairwise ΦST value estimated from the RFLP data, they exhibited a higher and significant pairwise ΦST value estimated from the sequence data, suggesting that Chança should also constitute an independent unit. These four isolated sets of populations may possess adaptations specific to local conditions, and conservation efforts should be directed towards preserving the genetic integrity of each group, because the failure to preserve distinctive stocks may reduce the evolutionary potential of the species. However, because they do not exhibit reciprocal monophyly, they cannot be considered as ESUs. As Moritz et al. (1995) pointed out, the definition of ESU does not take into consideration the potential contribution of stochastic lineage sorting in the initial differentiation of isolated populations.
The conservation units defined in the present study overlap, in general, the seven nuclei proposed by Collares-Pereira et al. (1999) (Fig. 1), which were defined taking into consideration recent physical barriers that may constrain migration. The only exceptions are the N1 (Caia) and N2 (Xévora) nuclei, which according to the present data constitute a single MU, and the Foupana River, which was included in N6 together with Carreiras and Vascão but constitutes an independent ESU. Consequently, N1 to N6 constitute a different ESU with four separate MUs, while Foupana (in N6) and Odeleite (N7) are considered independent ESUs.
The low to moderate dispersal ability of A. hispanica has important implications for its regional persistence. Migration seems to be low, even between some close populations (e.g. Chança vs. Carreiras or Vascão). In these cases, the chance of recolonization following an extinction event is correspondingly low. Low to moderate dispersal also limits the possibility of ‘topping up’ vulnerable populations, increasing their probability of extinction (reviewed in Hanski, 1999). These issues are particularly significant for the long-term persistence of A. hispanica, since the semiarid regime of the Guadiana River drainage together with the increasing human pressure make local bottlenecks and extinctions very likely, and may explain why this species is not found in some tributaries (e.g. Oeiras, Limas, Terges and Cobres), which apparently have suitable habitats for it (Collares-Pereira et al., 1999, 2000b).
References
Almaça, C. (1976). La speciation chez les Cyprinidae de la Péninsule Ibérique. Revue des Travaux de l’Inst des Pêches maritimes, 40: 399–411.
Almaça, C. (1995). Freshwater fish and their conservation in Portugal. Biol Conserv, 72: 125–127.
Avise, J. C. (2000). Phylogeography: The History and Formation of Species. Harvard University Press, Cambridge.
Avise, J. C., Neigel, J. E. and Arnold, J. (1984). Demographic influences on mitochondrial DNA lineage survivorship in animal populations. J Mol Evol, 20: 99–105.
Blanco, J. C. and González, J. L. eds.1992). Libro Rojo de los Vertebratos de España. Colección Técnica, Icona, Madrid. (In Spanish).
Brito, R. M., Briolay, J., Galtier, N. and Bouvet, Y. et al. (1997). Phylogenetic relationships within the genus Leuciscus (Pisces, Cyprinidae) in Portuguese fresh waters, based on mitochondrial DNA cytochrome b sequences. Mol Phylogenet Evol, 8: 435–442.
Chang, Y., Huang, F. and Lo, T. (1994). The complete nucleotide sequence and gene organization of carp (Cyprinus carpio) mitochondrial genome. J Mol Evol, 38: 138–155.
Collares-Pereira, M. J., Pires, A. M., Coelho, M. M. and Cowx, I. G. (1998). Towards a conservation strategy for Anaecypris hispanica, the most endangered non-migratory fish in Portuguese streams. In: Cowx, I. G. (ed.) Stocking and Introduction of Fish, pp. 437–449. Fishing News Books, Blackwell Science, Oxford.
Collares-Pereira, M. J., Cowx, I. G., Rodrigues, J. A. and Rogado, L. et al. (1999). The status of Anaecypris hispanica in Portugal: problems of conserving a highly endangered Iberian fish. Biol Conserv, 88: 207–212.
Collares-Pereira, M. J., Cowx, I. G., Ribeiro, F. and Rodrigues, J. A. et al. (2000a). Threats imposed by water development schemes on the conservation of endangered fish species in the Guadiana river basin in Portugal. Fish Manage Ecol, 7: 1–12.
Collares-Pereira, M. J., Rodrigues, J. A., Rogado, L. and Cowx, I. G. (2000b). Plano de Gestão para a Conservação do Saramugo. Programa Life Natureza, BA-3200/97/280, CBA-FCUL/ICN. (In Portuguese).
Cowx, I. G. and Collares-Pereira, M. J. (2000). Conservation of endangered fish species in the face of water resource development schemes in the Guadiana River, Portugal: harmony of the incompatible. In: Cowx, I. G. (ed.) Management and Ecology of River Fisheries, pp. 428–438. Fishing News Books, Blackwell Science, Oxford.
Crandall, K. A., Bininda-Emonds, O. R. P., Mace, G. M. and Wayne, R. K. (2000). Considering evolutionary processes in conservation biology. Trends Ecol Evol, 15: 290–295.
Dodson, J. J., Colombani, F. and Ng, P. K. L. (1995). Phylogeographic structure in mitochondrial DNA of a South-east Asian freshwater fish, Hemibagrus nemurus (Siluroidei: Bagridae) and Pleistocene sea-level changes on the Sunda shelf. Mol Ecol, 4: 331–346.
Elvira, B. (1995). Conservation status of endemic freshwater fish in Spain. Biol Conserv, 72: 129–136.
Excoffier, L. and Smouse, P. E. (1994). Using allele frequencies and geographic subdivision to reconstruct gene trees within a species: molecular variance parsimony. Genetics, 136: 343–359.
Excoffier, L., Smouse, P. E. and Quattro, J. M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics, 131: 479–491.
Gilles, A., Lecointre, G., Miquelis, A. and Loerstcher, M. et al. (2001). Partial combination applied to phylogeny of European cyprinids using the mitochondrial control region. Mol Phylogenet Evol, 19: 22–33.
Goldstein, P. Z., Desalle, R., Amato, G. and Vogler, A. P. (2000). Conservation genetics at the species boundary. Conserv Biol, 14: 120–131.
Hanski, I. (1999). Metapopulation Ecology. Oxford University Press, Oxford.
HIDROPROJECTO/COBA/HP (1998). Plano de Bacia Hidrográfica do Rio Guadiana. 1a Fase. Análise e Diagnóstico da Situação Actual. Anexo Temático 9. Conservação da Natureza. Parte 5. Análise da Hidrodinâmica do Estuário. Lisboa.
Hillis, D. M., Mable, B. K., Larson, A. and Davis, S. et al. (1996). Nucleic acids IV: Sequencing and cloning. In: Hillis, D. M., Moritz, C. and Mable, B. K. (eds) Molecular Systematics, 2nd edn, pp. 321–381. Sinauer, Sunderland, MA.
Hurwood, D. A. and Hughes, J. M. (1998). Phylogeography of the freshwater fish, Mogurnda adspersa, in streams of northeastern Queensland, Australia: evidence for altered drainage patterns. Mol Ecol, 7: 1507–1517.
INAG (INSTITUTO DA ÁGUA) (1996). Programa de Despoluição da Bacia do Rio Guadiana. (Versão de trabalho). Direcção de Serviços de Recursos Hídricos, INAG, Lisboa.
INAG (INSTITUTO DA ÁGUA) (1997). Melhoria e Controle da Qualidade da Água na Bacia do Rio Guadiana. (Versão preliminar), Direcção de Serviços de Recursos Hídricos, INAG, Lisboa.
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol, 16: 111–120.
Lande, R. and Barrowclough, G. F. (1987). Effective population size, genetic variation, and their use in population management. In: Soulé, M. E. (ed.) Viable Populations for Conservation, pp. 87–123. Cambridge University Press, Cambridge.
Maddison, W. P. and Maddison, D. R. (1992). MacClade, version 3.0., Sinauer, Sunderland, MA.
Mesquita, N., Carvalho, G., Shaw, P. and Crespo, E. et al. in press. River basin-related genetic structuring in an endangered fish species, Chondrostoma lusitanicum, based on mtDNA sequencing and RFLP analysis. Heredity, in press.
Moritz, C. (1994). Defining ‘Evolutionary Significant Units’ for conservation. Trends Ecol Evol, 9: 373–375.
Moritz, C., Lavery, S. and Slade, R. (1995). Using allele frequency and phylogeny to define units for conservation and management. In: Nielsen, J. (ed.) Evolution of the Aquatic Ecosystem. Defining Unique Units in Population Conservation. American Fisheries Society Symposium, 17, pp. 249–262.
Nei, M. (1987). Molecular Evolutionary Genetics. Columbia University Press, New York.
Nei, M. and Tajima, F. (1983). Maximum likelihood estimation of the number of nucleotide substitutions from restriction data. Genetics, 105: 207–217.
Neigel, J. E. and Avise, J. C. (1986). Phylogenetic relationships of mitochondrial DNA under various demographic models of speciation. In: Karvin, S. and Nevo, E. (eds) Evolutionary Processes and Theory, pp. 515–534. Academic Press, Orlando, FL.
Paetkau, D. (1999). Using genetics to identify intraspecific conservation units: a critique of current methods. Conserv Biol, 13: 1507–1509.
Ribeiro, F., Cowx, I. G. and Collares-Pereira, M. J. (2000). Life history traits of the endangered Iberian cyprinid Anaecypris hispanica and their implications for conservation. Arch Hydrobiol, 149: 569–586.
Schneider, S., Roessli, D. and Excoffier, L. (2000). Arlequin, version 2.000: A software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva. Switzerland.
Simberloff, D. (1988). The contribution of population and community biology to conservation science. Ann Rev Ecol Syst, 19: 437–511.
SNPRCN (ed.) (1991). Livro Vermelho dos Vertebrados, Vol. 2: Peixes Dulciaquícolas e Migradores. Secretaria de Estado dos Recursos Naturais, Lisboa. (In Portuguese).
Swofford, D. L. (2000). PAUP. Sinauer, Sunderland, MA.
Tamura, K. and Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol, 10: 512–526.
Tibbets, C. A. and Dowling, T. E. (1996). Effects of intrinsic and extrinsic factors on population fragmentation in three species of North American minnows (Teleostei: Cyprinidae). Evolution, 50: 1280–1292.
Weir, B. S. (1996). Intraspecific differentiation. In: Hillis, D. M., Moritz, C. and Mable, B. K. (eds) Molecular Systematics, 2nd edn, pp. 385–405. Sinauer, Sunderland, MA.
Acknowledgements
Thanks are especially due to J. A. Rodrigues, F. Ribeiro, L. Rogado, L. da Costa, P. Nunes, F. Filipe and T. Marques, who provided samples or supported fieldwork. We thank T. E. Dowling for the helpful discussions and comments on an early draft of the manuscript. The research was carried out under the project EU/LIFE-NATURE contract B4–3200/97/280.
Author information
Authors and Affiliations
Corresponding author
Appendix
Appendix
(See Appendix below)
Rights and permissions
About this article
Cite this article
Alves, M., Coelho, H., Collares-Pereira, M. et al. Mitochondrial DNA variation in the highly endangered cyprinid fish Anaecypris hispanica: importance for conservation. Heredity 87, 463–473 (2001). https://doi.org/10.1046/j.1365-2540.2001.00929.x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1046/j.1365-2540.2001.00929.x
Keywords
This article is cited by
-
Mitochondrial DNA revealed a single stock structure of the Spotted sardinella Amblygaster sirm (Walbaum, 1792) (Teleostei; Clupeidae) in Tanzanian coastal waters
Marine Biodiversity (2023)
-
Mitochondrial DNA based diversity studies reveal distinct and sub-structured populations of pearlspot, Etroplus suratensis (Bloch, 1790) in Indian waters
Journal of Genetics (2022)
-
The complete mitochondrial genome of an endangered minnow Aphyocypris lini (Cypriniformes: Xenocyprididae): genome characterization and phylogenetic consideration
Biologia (2021)
-
Cryptic lineages and hybridization of the predaceous chub Parazacco spilurus (Actinopterygii, Cypriniformes, Xenocyprididae) in Hong Kong
Hydrobiologia (2019)
-
Evolutionary history and population genetics of a cyprinid fish (Iberochondrostoma olisiponensis) endangered by introgression from a more abundant relative
Conservation Genetics (2014)