Abstract
A cytogenetical study was carried out on the chromosomes and nuclear DNA amounts of the terrestrial earthworms Octodrilus complanatus and Eisenia foetida (Annelida: Oligochaeta: Lumbricidae). Chromosomes were studied using Giemsa staining, banding methods and fluorescent in situ hybridization (FISH) with two repetitive DNA probes [rDNA and (TTAGGG)n]. rDNA FISH and silver staining consistently identified one chromosome pair per spread in both species. The telomeric sequence (TTAGGG)n hybridized with termini of all the chromosomes in both earthworms. Flow cytometry DNA assays showed that O. complanatus and E. foetida had different nuclear DNA contents (2C value=1.72 and=1.40 pg, respectively) but very similar base composition in their genomes.
Similar content being viewed by others
Introduction
FISH analysis with tandem highly repetitive DNAs [rDNA and (TTAGGG)n] proved to be very effective for understanding the organization of eukaryotic genomes at the molecular level (e.g. Trask, 1991; Phillips & Reed, 1996; and authors quoted by them). In particular, FISH with ribosomal sequences as a probe allowed the mapping of major rDNA clusters (18S, 5.8S and 28S) on the chromosomes in some taxonomically widely separated organisms, whose ribosomal sites remained unclear using conventional techniques (e.g. Pendás et al., 1993; Sánchez et al., 1995; Vitturi et al., 1999). Structural chromosomal rearrangements (i.e. para- and pericentric inversions and C–C or C–T Robertsonian translocations) were inferred from intrachromosomal location of the (TTAGGG)n telomeric sequence in some vertebrates (i.e. Meyne et al., 1990; Lee et al., 1993; Phillips & Reed, 1996).
Many vertebrates have been investigated with FISH but invertebrate taxa remain poorly characterized in this respect. One of the latter is the annelid class Oligochaeta, where the haploid (n) and/or the diploid (2n) chromosome number was determined for about 160 species from analysis of conventionally stained preparations (Christensen, 1980 and authors quoted by him).
A diploid chromosome number of 2n=36 was previously reported in O. complanatus (Muldal, 1952). In E. foetida a diploid set made up of 11 chromosome pairs of which five were metacentric/submetacentric (M/SM) and six subtelocentric (ST) was found by Vitturi et al. (1991). Silver staining and C-banding were also attempted on spermatogonial chromosomes of this species revealing argentophilic regions (Ag-NORs) terminally located on a single C-positive medium-sized chromosome pair (Vitturi et al., 1991). To the author’s knowledge, nuclear DNA contents have not been reported in Oligochaeta.
In the present work, major rDNA clusters were mapped and the presence of the telomeric repeat (TTAGGG)n was tested in the genome of E. foetida and O. complanatus. Further data, such as the description of a conventional karyogram and the detection of NORs in O. complanatus, along with nuclear DNA amounts in both species, were also collected.
Materials and methods
Octodrilus complanatus (Dugès 1828) and Eisenia foetida (Savigny 1826) are geographically widespread, terrestrial earthworms belonging to the family Lumbricidae (Annelida: Oligochaeta). The specimens used for the present study came from two different sites in the north-western part of Sicily (Palermo province, Italy) and were identified according to Bouché (1972). Chromosomes were prepared by means of the air-drying technique (Vitturi et al., 1991) from dissected testes of sexually mature animals previously soaked in fresh-water colchicine solution (0.1%) at room temperature (RT).
Conventional staining
Chromosome preparations were stained for 15 min in 5% Giemsa solution in phosphate buffer (pH 6.8) for conventional karyotyping, or left unstained for banding. Chromosomes were classified according to the criteria of Levan et al. (1964).
Banding techniques
Characterization of silver-stained Nucleolar Organizer Regions (Ag-NORs) was carried out using the one-step method (Howell & Black, 1980). Fluorescent staining with chromomycin A3 (CMA3) and 4′,6-diamidino-2-phenylindole (DAPI) were performed according to Schmid et al. (1983).
Probes and labelling
The telomeric probe was a digoxigenin-labelled all-human telomere probe (Oncor P4097-DG.5). The rDNA probe was a sea urchin (Paracentrotus lividus) 4.3 kilobase (kb) rDNA probe (prR14), consisting of sequences from the 3′ end of 18S rDNA to the 5′ end of 26S rDNA. Plasmid containing rDNA was labelled by nick translation with digoxigenin-11-dUTP according to manufacturer’s (Boehringer Mannheim) instructions.
In situ hybridization
Chromosomes were denatured for 4 min in 70% formamide/2 × SSC at 72°C. Denaturation of the probes was performed for 5 min at 80°C. Hybridization was allowed to proceed overnight at 37°C. Slides were washed twice in 50% formamide/2 × SSC at 40°C (5 min each), twice in 2 × SSC at 40°C (5 min each), once (5 min) in 4 × SSC/0.1% Tween at room temperature (RT) and finally, once (5 min) in PBS/0.1% Tween/0.5% skimmed milk powder at RT. Probe hybridization sites were detected using the ‘Fluorescent Antibody Enhancer Set for DIG Detection’ according to the manufacturer’s (Boehringer Mannheim) instructions. Slides were mounted in an antifade solution containing propidium iodide (5 μg/mL) and viewed under a Leica filter set I3 (BP 450–490, LP 515) which allowed the simultaneous visualization of fluorescein-labelled hybrid (yellow) and chromosomal DNA (red).
Chromosomes were observed with a Leica microscope and photographed on Agfaortho 25 ASA and Kodak Ektacolor 1000 ASA films.
Flow cytometry
The male gonads of the two earthworm species were also used as sources of cell suspensions for the evaluation of nuclear DNA content (DNA) and Adenine–Thymine base-pair nuclear DNA amount (AT-DNA) by flow cytometry. By mincing a frozen portion of the tissues with fine needles in phosphate saline buffer, a cell suspension was obtained. Each sample was divided into two subsamples: in one of them a proportion of chicken erythrocytes was added as an internal standard; the other subsample, containing only gonadal cells, was used in order to define the developmental stage of the testis and clearly identify the diploid peak position. The cells were stained with propidium iodide, after RNA digestion with RNAse, or with DAPI for DNA or AT-DNA determination, respectively. The DNA assays were performed on a BRYTE HS flow cytometer (Bio-Rad Laboratories Inc.) equipped with a Xenon–Mercury lamp. To determine DNA and AT-DNA, from nine to 14 samples for each species were analysed. At least 2500 worm cells per sample were considered. Diploid (2C) DNA and AT-DNA amounts of earthworms were calculated on the basis of the comparison with the internal standards, according to the equation W/C × S, where W and C are the respective mean fluorescence channel values of the diploid (2C) cell population of the worm and the chicken, and S=the DNA content (DNA or AT-DNA) of the standard (2C DNA=2.5 pg and 2C AT-DNA=1.39 pg: Tiersch & Chandler, 1989; Ronchetti et al., 1995). The two-tailed t-test was used to evaluate significance of the difference between the average values in the two species.
Results
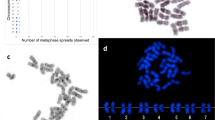
In the Sicilian strain of Octodrilus complanatus the diploid chromosome number was 2n=36. By arranging into pairs the chromosomes from 10 Giemsa-stained metaphase spreads according to dimension and arm ratio, a karyogram consisting of seven metacentric/submetacentric pairs (M/SM) (nos 5,6,7,13,15,16,18) and 11 subtelocentric pairs (ST) was produced (Fig. 1). After Ag-NOR staining, two small-sized chromosomes per spread were consistently found (10 specimens analysed) to bear argentophilic spots located subterminally on the short arm of a subtelocentric chromosome pair tentatively identified as no. 14 (Fig. 1, arrow). In the two species, both CMA3 and DAPI were uninformative for identifying NORs because all chromosomes showed a uniform dull fluorescence.
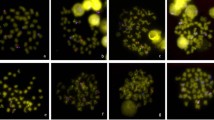
rDNA FISH was carried out on 10 specimens of each species (30 spreads per specimen analysed). Major rDNA cistrons were located terminally on two middle-sized submetacentrics in E. foetida (Fig. 2) and subterminally on the short arm of two small-sized subtelocentrics in O. complanatus (Fig. 2B), thus revealing a correspondence between rDNA mapping and NOR expression in the two species. Results with meiotic chromosomes confirmed the homology of rDNA FISH chromosomes because a single hybridized bivalent occurred in the corresponding spermatocytes.
Mitotic chromosomes after rDNA FISH treatment with a heterologous 18S rDNA probe from Paracentrotus lividus: E. foetida (A), O. complanatus (B). Spermatogonial chromosomes of E. foetida (C) and metaphase-I bivalents of O. complanatus (D) after FISH treatment with all-human telomeric sequence (TTAGGG)n.
Hybridization experiments with the (TTAGGG)n repeat, carried out on spermatogonial metaphase chromosomes of E. foetida (Fig. 2C), revealed small hybridization signals at each telomere of all the chromosomes of this species. A similar analysis carried out on spermatocyte metaphase-I chromosomes of O. complanatus showed small hybridized areas in all bivalents, presumably located in the telomeric regions (Fig. 2D).
Flow cytometrical analysis of the gonadal cell suspensions without chicken erythrocyte reference cells gave fluorescence histograms showing three peaks (corresponding to C, 2C and 4C DNA content nuclei) in mature testes, or two main peaks (2C and 4C) in immature or spent gonads. The fluorescence peak position of diploid (2C) cells for the two earthworm species was therefore determined.
Subsequently, the corresponding subsample with reference cells was run and the mean values of the diploid peak of the earthworm and the chicken (internal standard) were recorded. Estimated values of average diploid (2C) DNA content, AT-DNA amount and base composition (AT%) for the two earthworms are summarized in Table 1.
t-tests of the average values showed a significant difference between the two species (P < 0.001) for DNA and AT-DNA (t-values of 27.99 and 17.84 with 15 and 19 degrees of freedom, respectively). Ratios between E. foetida and O. complanatus average values were 0.81 and 0.79 for DNA and AT-DNA, respectively.
Discussion
This paper reports the first application of FISH to determine the physical location of repetitive DNAs [rDNA and (TTAGGG)n sequence] on chromosomes of Oligochaeta earthworms.
18S and 28S rDNA was consistently found on two homologous chromosomes per spread at spermatogonial metaphase both in Octodrilus complanatus and Eisenia foetida. In contrast to results in fish (e.g. Phillips & Hartley, 1988) and amphibian (e.g. Schmid et al., 1995), in the two earthworms we found that CMA3 proved inadequate to identify rDNA.
The presence of the (TTAGGG)n telomeric repeat was tested on E. foetida and O. complanatus chromosomes. Our results showed that the termini of each chromosome hybridized with the telomeric probe, revealing homology between the (TTAGGG)n all-human telomere sequence and the telomeres of the two earthworms. Similar homology was previously observed in two marine polychaete worms (Jha et al., 1995). Among nonvertebrate taxa, the presence of the (TTAGGG)n repeat has also been reported in trypanosomes (Van der Ploeg et al., 1984), slime moulds (Forney et al., 1987), the isopod crustacean Asellus aquaticus (Pelliccia et al., 1994) and the bulldog ants of the genus Myrmecia (Meyne et al., 1995).
O. complanatus and E. foetida karyotypes probably underwent extensive changes in chromosome number and morphology (because of chromosomal inversions and/or fusions) from the ancestral karyotype (Christensen, 1980; Vitturi et al., 1991) and there is no evidence of their mutual derivation. Results from the present study do not reveal the occurrence of (TTAGGG)n intrachromosomal signal in E. foetida chromosomes, while in O. complanatus, due to morphology of metaphase-I bivalents, this assessment cannot be conclusively demonstrated. The absence of interstitial telomeric signal in E. foetida may indicate that residual traces of inner telomeric sequences due to chromosomal rearrangements have been lost. In support to this assumption it may be pointed out that a prerequisite for Robertsonian fusion should be the elimination of telomeres (i.e. Garagna et al., 1995; Nanda et al., 1995; Slijepcevic, 1998) and that intrachromosomal blocks of (TTAGGG)n repeats are mainly conserved in species that underwent chromosome rearrangements during their recent evolution (Meyne et al., 1989) or in those showing intraspecific chromosome polymorphism (Reed & Phillips, 1995).
Although the in situ hybridization study with the (TTAGGG)n telomeric sequence gives no evidence of any chromosome rearrangement in E. foetida, available data from literature provide indirect proofs of its derived karyotypic condition. For example, a decrease of the chromosome number from the ancestral value 2n=36 (Christensen, 1980) to the derived one 2n=22, endowed by large-sized metacentric chromosomes, would indicate the occurrence of progressive Robertsonian translocations in this species (Vitturi et al., 1991).
Additional evidence to support a derived genomic condition in E. foetida, comes from the occurrence of a lower nuclear DNA content in this species than in O. complanatus: a species still conserving the ancestral diploid chromosome number (2n=36). Conversely, the finding of almost the same base composition in the two species would indicate that in E. foetida, loss of DNA was not accompanied by preferential changes of AT- or GC-fractions.
References
Bouché, M. B. (1972). Lombriciens de France. Écologie et systématique. Institut National de la Recherche Agronomique (ed), 72: 311–387.
Christensen, B. (1980). Animal Cytogenetics. B. John (ed.), vol. 2. Borntraeger, Stuttgart.
Forney, Y. J., Henderson, E. R. and Blackburn, E. H. (1987). Identification of the telomeric sequence of the acellular slime moulds Didimium iridis and Physarum polycephalum. Nucl Acids Res, 15: 9143–9152.
Garagna, S., Broccoli, D., Redi, C. A., Searle, J. B., Cooke, H. J. and Capanna, E. (1995). Robertsonian metacentrics of the mouse lose telomeric sequences but retain some minor satellite DNA in the pericentric area. Chromosoma, 103: 685–692.
Howell, W. M. and Black, D. A. (1980). Controlled silver-staining of nucleolus organizer regions with a protective colloidal developer: a 1-step method. Experientia, 36: 1014–1014.
Jha, A. N., Dominquez, I., Balajee, A. S., Hutchinson, T. H., Dixon, D. R. and Natarajan, A. T. (1995). Localization of a vertebrate telomeric sequence in the chromosomes of two marine worms (phylum Annelida: class Polychaeta). Chrom Res, 3: 507–508.
Lee, C., Sasi, R. and Lin, C. C. (1993). Interstitial localization of telomeric DNA sequences in the Indian muntjac chromosomes: further evidence for tandem chromosome fusions in the karyotype evolution of the Asian muntjacs. Cytogenet Cell Genet, 63: 156–159.
Levan, A., Fredga, K. and Sandberg, A. A. (1964). Nomenclature for centromeric position of chromosomes. Hereditas, 52: 201–220.
Meyne, J., Ratcliff, L. R. and Moyzis, R. K. (1989). Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc Natl Acad Sci USA, 86: 7049–7053.
Meyne, J., Baker, R. J., Hobart, H. H., Hsu, T. C., Oliver, A. R., Ward, O. G. et al (1990). Distribution of non-telomeric sites of the (TTAGGG)n telomeric sequences in vertebrate chromosomes. Chromosoma, 99: 3–10.
Meyne, J., Hirai, H. and Imai, H. T. (1995). FISH analysis of the telomere sequences of bulldog ants (Myrmecia: Formicidae). Chromosoma, 104: 14–18.
Muldal, S. (1952). The chromosomes of the earthworms. I. The evolution of polyploidy. Heredity, 6: 55–76.
Nanda, I., Schneider-Rasp, S., Winking, H. and Schmid, M. (1995). Loss of telomeric sites in the chromosomes of Mus musculus domesticus (Rodentia: Muridae) during Robertsonian rearrangements. Chrom Res, 3: 399–409.
Pelliccia, F., Volpi, E. V., Lanza, V., Gaddini, L., Baldini, A. and Rocchi, A. (1994). Telomeric sequences of Asellus aquaticus (Crust. Isop.). Heredity, 72: 78–80.
Pendás, A. M., Morón, P. and Garcia-Vázquez, E. (1993). Multi-chromosomal location of ribosomal RNA genes and heterochromatin association in brown trout. Chrom Res, 1: 63–67.
Phillips, R. B. and Hartley, S. E. (1988). Fluorescent banding patterns of the chromosomes of the genus Salmo. Genome, 30: 193–197.
Phillips, R. B. and Reed, K. M. (1996). Application of fluorescence in situ hybridization (FISH) techniques to fish genetics: a review. Aquaculture, 140: 197–216.
Reed, K. M. and Phillips, R. B. (1995). Molecular cytogenetic analysis of the double-CMA3 chromosome of lake trout, Salvelinus namaycush. Cytogenet Cell Genet, 70: 104–107.
Ronchetti, E., Salvadori, S. and Deiana, A. M. (1995). Genome size and AT-content in Anguilliformes. Europ J Histochem, 39: 259–264.
Sánchez, A., Jímenez, R., Burgos, M., Stitou, S., Zurita, F. and Díaz de la Guardia, R. (1995). Cytogenetic peculiarities in the Algerian hedgehog: silver stains not only NORs but also heterochromatic blocks. Heredity, 75: 10–16.
Schmid, M., Haaf, T., Geile, B. and Sims, S. (1983). Chromosome banding in Amphibia. VIII. an unusual XY/XX sex chromosome system in Gastrotheca riobambae (Anura, Hylidae). Chromosoma, 88: 69–82.
Schmid, M., Feichtinger, W., Weimer, R., Mais, C., Bolaños, F. and León, P. (1995). Chromosome banding in Amphibia. XXI. Inversion polymorphism and multiple nucleolus organizer regions in Agalychnis callidryas (Anura, Hylidae). Cytogenet Cell Genet, 69: 18–26.
Slijepcevic, P. (1998). Telomeres and mechanisms of Robertsonian fusion. Chromosoma, 107: 136–140.
Tiersch, T. R. and Chandler, R. W. (1989). Chicken erythrocytes as an internal reference for analysis of DNA content by flow cytometry in the Carp. Trans Am Fish Soc, 118: 713–717.
Trask, B. (1991). Fluorescence in situ hybridization: application in cytogenetics and gene mapping. Trends Genet, 7: 149–154.
van Der Ploeg, L. H. T., Lin, A. Y. C. and Borst, P. (1984). Structure of the growing telomeres of Trypanosomes. Cell, 36: 459–468.
Vitturi, R., Colombera, D., Catalano, E. and Amico, F. P. (1991). Karyotype analysis, nucleolus organizer regions and C-banding pattern of Eisenia foetida (Oligochaeta, Lumbricidae). Genetica, 83: 159–165.
Vitturi, R., Colomba, M. S., Barbieri, R. and Zunino, M. (1999). Ribosomal DNA location in the scarab beetle Thorectes intermedius (Costa) (Coleoptera: Geotrupidae) using banding and fluorescent in situ hybridization. Chrom Res, 7: 255–260.
Acknowledgements
This work was supported by a MURST grant (ex 60%) to R. Vitturi. The ‘Azienda Multiservizi Ambientali Veneziana’ of Venice (Italy) is thanked for providing the flow cytometer.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Vitturi, R., Colomba, M., Pirrone, A. et al. Physical mapping of rDNA genes, (TTAGGG)n telomeric sequence and other karyological features in two earthworms of the family Lumbricidae (Annelida: Oligochaeta). Heredity 85, 203–207 (2000). https://doi.org/10.1046/j.1365-2540.2000.00709.x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1046/j.1365-2540.2000.00709.x
Keywords
This article is cited by
-
Telomere maintenance during anterior regeneration and aging in the freshwater annelid Aeolosoma viride
Scientific Reports (2018)
-
Alike but different: the evolution of the Tubifex tubifex species complex (Annelida, Clitellata) through polyploidization
BMC Evolutionary Biology (2014)
-
Chromosome number and telomere sequence mapping of the Japanese sea cucumber Apostichopus japonicus
Fisheries Science (2009)