Abstract
Introduced populations of weeds which are polyploid and reproduce primarily by apomixis are generally considered as having low levels of population genetic variation, highly differentiated populations and short evolutionary lifespans. Although polyploidy allows for habitat differentiation and colonization, lack of recombination because of apomixis means that long-term persistence is unlikely. However, variation can be introduced to a colonizing population by evolutionary changes in the mating system, or by somatic mutation and recombination. In this study hypersensitive genetic markers, inter-simple sequence repeats (ISSRs), were used to quantify genetic variation within Pilosella officinarum, a major weed of the New Zealand high country. Pilosella officinarum was introduced from Europe to New Zealand late in the 19th century and only polyploid, apomictic populations are thought to have survived. The combination of introduction history and breeding system has led to the assumption that New Zealand populations are necessarily genetically depauperate. However, our studies reveal variable levels of genetic variation and patterns of clonal distribution which indicate varying levels of sexual reproduction within New Zealand populations.
Similar content being viewed by others
Introduction
There is now a considerable literature describing the general characteristics of invasive plant species (Thomson, 1991; and see Roy, 1990 for general review), but characteristic patterns of genetic variation associated with successful colonizers have not been identified (Roy, 1990; Schierenbeck et al., 1995). However, there is general agreement in the literature that adventive populations of clonal species show low levels of genetic variation compared with native populations because of breeding system and founder effects (Brown & Marshall, 1981; Barrett, 1992). Intuitively this makes sense; clonal reproduction in plants is manifested as either vegetative propagation or agamospermous apomixis (asexual reproduction through seed). Apomictic seed is produced without meiosis or fertilization, so that in the absence of somatic mutations, the offspring have the genetic constitution of the mother plant (Nogler, 1984; Calzada et al., 1996). Indeed, early population biologists, such as Darlington (1939) (but see Richards, 1996) and Mayr (1970), considered apomictic species as ‘evolutionary dead-ends’. Such a breeding system, in conjunction with founder events, must result in reduced levels of genetic variation. However, Ellstrand & Roose’s (1987) review of genotypic diversity in clonal plants challenged these assumptions. Based only on morphological and isozyme studies, the review showed that most predominantly asexual populations had intermediate, not necessarily low, levels of diversity. Furthermore, recent empirical studies using various DNA markers have shed further doubt on the generally accepted adage that asexual populations are necessarily less genetically diverse than sexual ones (e.g. Diggle et al., 1998; and see Widén et al., 1994 for a review). Nevertheless, all adventive colonizers must undergo one or several founder events, and the breeding system should, theoretically, determine the rate at which variation is regenerated. Theory predicts that it will be slower in clonal and inbreeding populations than in outcrossing ones.
Despite the development of theoretical models for estimating the degree of sexuality in facultative apomicts (Barrett & Richardson, 1986), relatively little is known of their reproductive biology in natural populations (Barrett, 1992).
Here we determine levels of genetic variation within and between populations of an extremely successful adventive, facultative apomictic species in order to investigate whether success is facilitated by the generation of genetic variation through either sexual reproduction, mutation or a combination of both.
Pilosella officinarum (F.W. Schultz & Sch. Bip.) (Hieracium pilosella L.), native to Britain and Europe, is an extremely successful colonizer (sensu Thomson, 1991) in New Zealand, where it was accidentally introduced in the late 19th century as a contaminant of grass seed. It has since colonized a wide geographical area, exists over a range of localized environmental conditions and forms a dominant component of the vegetation into which it spreads (Scott, 1984, 1993; Hunter, 1991).
Pilosella officinarum has a complex reproductive strategy. It reproduces vegetatively through ramet fragmentation and by the production of both sexual and aposporous apomictic seed. That is, in the developing ovule of apomictic biotypes the products of meiosis are displaced, and typically destroyed, by one or more embryo sacs arising directly from the somatic cells of the nucellus (Asker & Jerling, 1992). As a result, at anthesis most ovules contain only unreduced or ‘maternal’ embryo sacs. In Europe both sexual diploid and polyploid aposporic apomict plants are common, so that European populations typically have a complex population structure because of hybridization, polyploidy and reproduction by apomixis (Gadella, 1991).
In New Zealand all P. officinarum populations appear to comprise only polyploid, facultatively apomictic plants (Makepeace, 1981; Jenkins & Jong, 1997). Recent experiments have shown that approximately 1% of seed produced under glasshouse conditions is sexual (H.M. Chapman & Bicknell, unpubl. data).
This ecology, combined with breeding system and history of introduction, makes New Zealand populations of P. officinarum ideal for testing the hypothesis that clonal facultatively apomictic populations comprise low levels of genetic variation.
Materials and methods
During the spring of 1994, 50 ramets from individual clones of P. officinarum were collected from each of five sites in the South Island of New Zealand: Cave Stream, Tekapo, Twizel, Halden and Hinewai (Fig. 1). Sites were chosen to include both hierarchical clustering of geographical distance and different histories of colonization by P. officinarum. Tekapo, Twizel and Halden, which comprise the ‘Mackenzie sites’, are within 50 km of each other and have a long history of P. officinarum colonization, with it being recognized as a common, weedy species in these areas by the end of the Second World War (Scott, 1993). Hinewai is approximately 250 km north north-east of the Mackenzie sites. Although P. officinarum was common here during the 1980s, it has since experienced a severe population contraction; there were thousands of ramets but there are now fewer than a hundred. Cave Stream is approximately 200 km north-east of the Mackenzie sites and 120 km north-west of Hinewai. Pilosella officinarum has been a weedy species here since the 1950s. Characteristics of the sites are summarized in Table 1.
To ensure that no individuals were of the same clone, P. officinarum ramets were sampled at sites greater than 5 m apart. Ramets were propagated and transferred to uniform conditions in a glasshouse at the University of Canterbury, Christchurch, New Zealand. Pots were laid out in a random block design in the glasshouse, and were subsequently split and repotted on a regular basis to reduce overcrowding and promote flowering.
Thirty ramets were selected for further analysis from each of the five sites, giving a total sample size of 150 ramets.
Root tips were collected from actively growing plants maintained in the glasshouse on a bed of moist sand. The tissue was treated for 3 h in a saturated aqueous solution of para-dichlorobenzene and 8-hydroxyquinoline, fixed in 3:1 ethanol:glacial acetic acid, hydrolysed for 15 min in 1 M HCl at 60°C, then stained for 2 h in a 1% solution of aceto-orcein. After a brief period of destaining in 45% acetic acid, the tissue was gently compressed, then mounted in 45% acetic acid and viewed under oil immersion. Only cells with an intact cytoplasm and well-separated chromosomes were used to assess ploidy level (Lambie, 1999).
Because of the clonal nature of P. officinarum, it was decided to use highly informative genetic markers for this study: 5′-anchored intersimple sequence repeat (ISSR) primers, which are abundant throughout the eukaryotic genome and target highly variable and numerous loci (Zietkiewicz et al., 1994), were chosen. They have a proven record in the genotyping of cultivated plants (Charters et al., 1996; Fang & Roose, 1997) and have been shown to be more sensitive than RAPDs as a means of characterizing complex genomes (Charters et al., 1996). Wolfe et al. (1998) list the advantages of ISSRs over other fingerprinting techniques.
Fresh leaf tissues were used for total genomic DNA isolation. Leaf tissue (0.10–0.15 g) was crushed in a mortar containing 20–40 mg of polyvinyl polypyr-rolidone (PVPP-40) and 500 μL of isolation buffer (200 mM Tris-HCl (pH 8.0), 250 mM NaCl, 25 mM EDTA, 0.5% sodium dodecyl sulphate (SDS) and 10 mM β-mercaptoethanol). The mixture was transferred to a 1.5-mL microfuge tube and incubated at 37 °C for 3 h, then centrifuged at 11 000 g for 5 min. The resulting supernatant was transferred to a clean microfuge tube and washed twice with phenol:chloroform:iso-amyl alcohol (25:24:1 v/v) and once with chloroform:iso-amyl alcohol (24:1 v/v). At each wash, the mixture was centrifuged for 5 min at 8000 g. The aqueous phase was transferred to a clean microfuge tube to which 300 μL of cold isopropanol was added. The mixture was incubated at −20°C for 20 min to allow DNA precipitation. The DNA was pelleted by centrifuging at 11 000 g for 5 min, washed with 70% ethanol, spun at 8000 g for 3 min, air-dried for 3 min and re-suspended in 50 μL of TE (tris acetate; 10 mM Tris-HCl (pH 8.0), 1 mM EDTA). The DNA was incubated at 37 °C for 30 min following addition of 5 μL of RNAse (10 μg μL–1) and re-precipitated with cold isopropanol. After an ethanol wash, the DNA was centrifuged at 8000 g for 3 min, air-dried and resuspended in an appropriate amount of TE.
The DNA was quantified by measuring the absorbance in a spectrophotometer at 260 nm.
DNA amplification and electrophoresis
The ISSR primers were supplied by the University of British Columbia Biotechnology Laboratory as primer set 9, and ISSRs were amplified by the modified PCR procedure of Williams et al. (1990). One hundred ISSR primers were screened over 20 samples. PCR was performed in a 50 μL reaction mixture per sample (1 × Taq polymerase PCR buffer, 400 μM dNTPs, 6 mM magnesium chloride, 0.2 μM of primer, 2.5 U of Taq DNA polymerase (Qiagen) and 100 ng of genomic (DNA). This amplification was carried out in a PTC-200 Thermal Cycler (MJ Research). Initial denaturation was for 4 min at 94 °C, followed by 45 cycles of 9 s at 94 °C, 27 s at 36 °C, 20 s at 72 °C, and a final extension of 4 min at 72 °C. The PCR products were separated electrophoretically on 2% agarose gels in 1 × TBE buffer and stained with ethidium bromide. The presence or absence of bands was scored under UV illumination. Six primers (Table 2) that gave clear and consistent banding patterns were selected for the analysis of the complete sample set.
Bands were scored based on their reproducibility and consistency in order to determine the ISSR phenotype for each individual sampled. Only bands which were clear and reproducible were included in the analysis. Each reaction was repeated three times to confirm reproducibility.
The Nei & Li (1979) similarity index (S ) was calculated to compare the presence/absence of ISSR fragments: S=2mxy/(mx + my), where mxy is the number of bands shared by the two patterns x and y, mx is the total number of bands in pattern x and my is the total number of bands in pattern y. Using this method, no assumptions had to be made regarding the number of loci or alleles. A distance matrix (D=1 − S ) was calculated using the software package RAPDistance version 1.04 (Armstrong et al., 1996). Both neighbour-joining and UPGMA trees were constructed using RAPDistance and the software package PHYLIP version 3.5c (Felstenstein, 1995), and MVSP version 3.1 (Kovach Computing Services Pentraeth, UK 1998), respectively.
Principal coordinate analysis (PCO) of the distance matrix followed Mardia et al. (1979) using the statistical package NTSYS-pc, version 2 (Applied Biostatistics Inc. 1990, USA).
ISSR-PCR does not provide complete genotypic information, and therefore genetic diversity statistics such as Wright’s F-statistics cannot be used for analysis. Although Lynch & Milligan (1994) provide a method for estimating allele frequencies using the frequency of the null allele, clonal reproduction makes deviations from Hardy–Weinberg equilibrium very likely in these populations, so their method is not applicable. An estimate of clonal diversity was obtained using the Simpson diversity index (D). As D depends on the sample size, the Simpson index of evenness (J ) was also used to illustrate the evenness of occurrence of clones. This analysis was performed using the program MVSP Plus version 3.1. In addition, the mean within-site interclonal similarity was calculated. This measure is the mean genetic distance between all clones present at a site, and provides an indication of the relationship between clones independent of their frequency.
Results
Table 3 shows that the majority of the 50 (out of the total 150) individual ramets sampled were either pentaploid (2n=45; 60%), tetraploid (2n=36; 14%), or aneusomatic tetraploid (chromosome numbers within the same ramet varied between 39 and 46; 22%). There were also 2% of 2n=63 and 2n=72 types.
The analysis produced a total of 32 ISSR bands, of which 16 (50%) were polymorphic (Table 4). Of these 16 polymorphic loci, nine were fixed in at least one of the five populations. Where they were not fixed the frequency of these markers ranged between 43 and 97%, although most were scored with a high frequency of between 87 and 97%. Seven of the polymorphic loci were not fixed at any of the sites. These only occurred at relatively low frequencies, which ranged from only 3 to 33%.
Primer 866 produced the highest frequency (83%) of polymorphic bands. Five of the six bands were found in each of the five populations, but with frequencies ranging from 43 to 100%. Band 866_6 was rare, and was only recorded from a single clone growing at Cave Stream and Twizel. This was a common pattern; six out of the nine bands produced from primer 895 were recorded from all five populations. The three ‘primer 895’ bands, restricted to just a few populations, were only recorded at low frequency; for instance, band 895_4 was scored from a rare clone from Twizel, and 895_7 from one and three individuals from Tekapo and Hinewai, respectively. Only one band, 895_4, was unique to a single population, and this was recorded from a single ramet from Twizel.
A total of 39 ISSR phenotypes (clones) were identified from the five sites. Thirteen of these clones comprised >1 ramet, and have been named clones A–M (Table 5). The remaining 26 clones were only represented by a single ramet, and are hereafter referred to as ‘rare clones’.
Clones A–M accounted for only 33.3% of clonal diversity (Table 5). Clone A was the most common clone, comprising 68 of the 150 individuals (45%) of all the ramets sampled. It was identified at all five sites, as was the second most abundant, clone B. Clone B, however, comprised only 14 (9.4%) of all ramets. The remaining 11 clones (C–M), were even less common, comprising between 10 (6.7%) and 2 (1.3%) of all the ramets sampled. These clones were either scattered among the sites, or restricted to a specific site, with no obvious correlation with the environment (Table 5). In contrast the 26 ‘rare clones’ comprised 66.7% of the total clonal diversity. There were far more rare clones at Twizel (17) than at any of the other sites (1–4).
Cave Stream, Tekapo, Halden and Hinewai all shared a similar pattern of clonal diversity. They all comprised between four (Tekapo) to seven (Cave Stream) clones and one (Halden) to four (Hinewai) ‘rare clones’ (Table 5).
Clone A, the most common clone, comprised between 23.3% (Cave Stream) and 34.3% (Halden) of the ramets sampled from these four sites. Clone B always comprised either 29% or 14% (Table 5). The remaining clones were distributed among the sites with no obvious pattern to their distribution. Clones C–H were found in more than one site, whereas clones I–M and all the ‘rare clones’ (by definition) were recorded from only one site.
Twizel differed from the other four sites; it had a relatively high level of clonal diversity in both clones and ‘rare clones’. Eight clones and 17 ‘rare clones’ were recorded at Twizel, four times as many as from any of the other sites.
These patterns are reflected in the equitability values (J), which range from 0.489 to 0.988, and in the Simpson Diversity Index (D), which ranges from 0.391 to 0.949 (Table 5).
Although within-site clonal diversity varied considerably among sites, genetic distances among clones within sites did not. This was to be expected, as the number of variable bands within sites was low, ranging from three to seven, except for Twizel with 13 variable bands (Table 4). The mean interclonal similarity coefficients of all sites except for Twizel exceeded 0.96 and also had low standard deviations, indicating high levels of within-site genetic relatedness (Table 5).
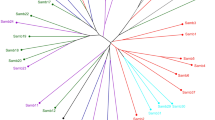
Both principal coordinate analysis (Fig. 2) and cluster analysis (Fig. 3) group the ramets into 13 clones and 26 ‘rare clones’ distributed among sites. The similarity coefficients among clones at different sites were generally less than those among clones at the same site, but even these were high, ranging between 0.83 and 1.
Both the neighbour-joining and UPGMA dendrograms assigned ramets to the same clone with a high degree of certainty. In fact, the clonal distribution was almost identical using a range of similarity/distance coefficients as well as tree building techniques. Distances among clusters were relatively uniform and did not resemble the spatial arrangement of the sampling sites. A Mantel test indicated no significant relationship between genetic distance and geographical distances (R2=0.0111).
Discussion
Successful colonizing species will have experienced repeated founder events, and are therefore likely to show widely different clonal structure and genetic diversity in different parts of the geographical range (Barrett, 1992). In addition, it is generally accepted that species undergoing bottlenecks or founder events will show low levels of genetic variation relative to conspecifics (Barrett, 1992; Schierenbeck et al., 1995). Furthermore, clonal weeds such as apomicts are expected to take a longer time to recover genetic variation after a founder event than sexually reproducing outcrossers (Barrett, 1992). These assumptions have been behind the rationale that host-specific pathogens will be more successful as biocontrol agents on clonal than outcrossing species (Barrett, 1992). The results of this study shed some doubt on these generalizations.
It has not been possible to compare the genetic diversity of New Zealand populations of P. officinarum with those of European populations because there have been no population-level molecular studies carried out on the latter. Instead, a comparison was made between our results for clonal diversity in P. officinarum and levels of clonal diversity recorded for other species of Asteraceae. In their review of clonal genotypic diversity (based primarily on allozyme, but including RAPD analyses), Widén et al. (1994) summarized diversity data from 30 plant species, and included: mean number of genotypes per population and range; total number of genotypes per study; percentage of genotypes found in only one population; and Simpson’s index of clonal diversity. When our data are compared with those for the eight apomict Asteraceae included in the review (Table 6), it is clear that New Zealand P. officinarum is not unusual in its amount and patterns of genetic diversity. Mean number of genotypes per population is lower than that reported for Erigeron annus, but higher than Antennaria rosea and five of the six species of Taraxacum listed (Table 6). Likewise, Simpson’s index of diversity was slightly lower for P. officinarum than for E. annus but higher than calculated values for T. officinale and T. tortilobum. The main difference between P. officinarum and the other species included in Table 6 lies in the distribution of widespread genotypes. There are fewer widespread genotypes (those genotypes occurring in more than three-quarters of all the populations) recorded for P. officinarum, than all the other asteraceous species included in the review, except for the ‘obligate’ apomicts, A. rosea and T. vindobonense. Of course, these comparisons are limited as we are comparing data sets of different sizes, allozyme data with ISSR data, and apomicts with different levels of sexuality. However, the comparisons do suggest that P. officinarum is not unusual in its clonal diversity. Indeed, Twizel’s pattern of genotypic diversity is typical of clonal populations: a high number of genotypes per population but the mean number of populations in which a given genotype occurs being restricted (Ellstrand & Roose, 1987).
But why has Twizel such high levels of diversity compared to the other four New Zealand populations? Further work is needed to answer this question, but it does indicate that clonal, polyploid populations do have the ability rapidly to regain variation which may be lost through founder events. One explanation which warrants investigation is that of higher levels of sexual reproduction at Twizel compared to the other four sites. Approximately 1% of P. officinarum seed from these populations has already been shown to be sexual under glasshouse conditions (H.M. Chapman & R.A. Bicknell, unpublished data) and it may involve only a minor physiological change to produce primarily sexual rather than apomictic seed. Alternatively there may be similar levels of sexual reproduction at all sites, but strongest selection for sexual plants at Twizel. A further possibility, which is again dependent upon segregation and recombination, is that current levels of clonal diversity reflect ‘frozen’ variation; that is variation which arose through sexual reproduction in an ancestral population but has since become fixed through polyploidy and apomixis (J.C.M. den Nijs, personal communication). Somatic mutation may explain some of the unique clones, especially those which vary from others by a single additional band.
Introduction and management history may be important in determining the structure of genetic variation. For all five sites there is strong evidence to suggest that the P. officinarum populations at each site originated from the same seed source. This probably reflects the fact that the pasture grass seed (of which P. officinarum was a contaminant) sown in the three regions came from the same European source. Alternatively, subsequent spread from a common New Zealand source, combined with selection and drift, may explain the observed pattern of clonal distribution. Pilosella officinarum has been regarded as a weedy species in the Mackenzie country and North Canterbury since the late 1940s (Scott, 1984), and since the early 1970s the population has expanded rapidly (Scott, 1993). At Hinewai, however, it was not recognized as a major weedy species until the late 1960s and since then has undergone a reduction in population size resulting from a change in management from heavy to lax grazing.
In contrast to European populations of P. officinarum, New Zealand populations are entirely polyploid, and tetraploids at least are capable of sexual reproduction in the field (G. Houliston & H.M. Chapman, unpublished data). These attributes confer on a species the potential to colonize new habitats, evolve rapidly and persist (Lumaret et al., 1987; Thomson & Lumaret, 1992). The fact that Makepeace (1981) recorded only pentaploids from all his New Zealand sites suggests that perhaps new polyploid races have evolved within New Zealand. This may explain why, in New Zealand, P. officinarum is such a successful invasive species. Breeding system, polyploidy and perhaps somatic recombination, all contribute to its success. They also ensure that any host-specific pathogen may be ineffectual as a biocontrol agent, as P. officinarum may be able to evolve resistance rapidly. Indeed, it may explain why the rust fungus Puccinia hieracii var. piloselloidarum, currently used as a biocontrol agent, is so variable in its efficacy (Morin & Syrett, 1996).
References
Armstrong, J., Gibbs, A., Peakall, R. and Weiller, G. (1996) The RAPDistance package, Version 1.04. http://life.anu.edu.au/molecular/software/rapd.htm.
Asker, S. E. and Jerling, L. (1992) Apomixis in Plants. CRC Press, Boca Raton, FL.
Barrett, S. C. H. (1992). Genetics of weed invasions. In: Jain, S. K. and Botsford, L. W. (eds) Applied Population Biology, pp. 91–119. Kluwer Academic Publishers, The Netherlands.
Barrett, S. C. H. and Richardson, B. J. (1986). Genetic attributes of invading species. In: Groves, R. H. and Burdon, J. J. (eds) Ecology of Biological Invasions, pp. 21–33. Cambridge University Press, Melbourne.
Brown, A. D. H. and Marshall, D. R. (1981). Evolutionary changes accompanying colonization in plants. In: Scudder, G. E. and Reveal, J. L. (eds) Colonization, Succession and Stability, pp. 351–363. Blackwell Scientific Publications, Oxford.
Calzada, J. P. V., Crane, C. F. and Stelly, D. M. (1996). Apomixis: the asexual revolution. Science. 274: 1322–1323.
Charters, Y. M., Robertson, A., Wilkinson, M. J. and Ramsey, G. (1996). PCR analysis of oilseed rape cultivars (Brassica napus L. ssp. olifera) using 5′-anchored simple sequence repeat (SSR) primers. Theor Appl Genet. 92: 442–447.
Darlington, C. D. (1939) The Evolution of Genetic Systems. Oliver and Boyd, Edinburgh.
Diggle, P. K., Lower, S. and Ranker, T. A. (1998). Clonal diversity in alpine populations of Polygonum viviparum (Polygonaceae). Int J Plant Sci. 159: 606–615.
Ellstrand, N. C. and Roose, M. L. (1987). Patterns of genotypic diversity in clonal plant species. Am J Bot. 74: 123–131.
Fang, D. Q. and Roose, M. L. (1997). Identification of closely related citrus cultivars with inter-simple sequence repeat markers. Theor Appl Genet. 95: 408–417.
Felstenstein, J. (1995) Phylip (Phylogeny Inference Package), Version 3.5c. Department of Genetics, University of Washington, Seattle, WA.
Floate, M. and Cossens, G. (1992). Land Resources. In: Floate, M. (ed.) Guide to Tussock Grassland Farming, pp. 25–33. AgResearch, Dunedin.
Gadella, T. W. J. (1991). Variation, hybridization and reproductive biology of Hieracium officinarum L. Proc Kon Ned Akad v Wetensch. 94: 455–488.
Hunter, G. (1991). The distribution of Hawkweeds (Hieracium) in the South Island, indicting problem status. Tuss Grass & Mount Land Inst Rev. 48: 1–10.
Jenkins, T. A. and Jong, K. (1997). Significance of polyploid variation in New Zealand Officinarum and Hieracium (Asteraceae). Bot J Scot. 49: 75–88.
Lambie, S. (1999). The role of retrostransposons in the evolution of Hieracium. MSc Thesis, University of Canterbury, New Zealand.
Lumaret, R., Guillerm, L., Delay, J., Ait, L. H. A. J., Loutfi, A., Izco, J. and Jay, M. (1987). Polyploidy and habitat differentiation in Dactylis glomerata L. from Galicia (Spain). Oecologia. 73: 436–446.
Lynch, M. and Milligan, B. G. (1994). Analysis of population genetic structure with RAPD markers. Mol Ecol. 3: 91–99.
Makepeace, W. (1981). Polymorphism and the chromosomal number of Hieracium officinarum L. in New Zealand. N Z J Bot. 19: 255–257.
Mardia, K. V., Kent, J. T. and Bibby, J. M. (1979) Multivariate Analysis. Academic Press, London.
Mayr, E. (1970) Populations, Species and Evolution. Belknap, Cambridge, MA.
Morin, L. and Syrett, P. (1996) Prospects for biological control of Hieracium pilosella with the rust Puccinia hieracii var. piloselloidarum in New Zealand. Proceedings of the IX International Symposium on Biological Control of Weeds, pp. 199–204. Stellenbosch, South Africa.
Nei, M. and Li, W. H. (1979). Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA. 76: 5269–5273.
Nogler, G. A. (1984). Gametophytic apomixis. In: Johri, B. M. (ed.) Embryology of Angiosperms, pp. 475–518. Springer-Verlag, Berlin.
Richards, A. J. (1996). Genetic variability in obligate apomicts of the genus Taraxacum. Fol Geo Phyto. 31: 405–414.
Roy, J. (1990). In search of the characteristics of plant invaders. In: Di Castri, F., Hansen, A. J. and Debussche, M. (eds) Biological Invasions in Europe and the Mediterranean Basin, pp. 335–352. Kluwer Academic Publishers, London.
Schierenbeck, K. A., Hamrick, J. L. and Mack, R. N. (1995). Comparison of allozyme variability in a native and an introduced species of Lonicera. Heredity. 75: 1–9.
Scott, D. (1984). Hawkweeds in run country. Tuss Grass & Mount Land Inst Rev. 42: 33–48.
Scott, D. (1993). Time segment analysis of permanent quadrat data: changes in Hieracium cover in the Waimakariri in 35 years. N Z J Ecol. 17: 53–57.
Thomson, J. D. (1991). The biology of an invasive plant. What makes Spartina anglica so successful? Bioscience. 41: 393–401.
Thomson, J. D. and Lumaret, R. (1992). The evolutionary dynamics of polyploid plants: origins, establishment and persistence. Trends Ecol Evol. 7: 303–307.
Widén, B., Cronberg, N. and Widén, M. (1994). Genotypic diversity, molecular markers and spatial distribution of genets in clonal plants, a literature survey. Folia Geobot Phytotax, Praha. 29: 245–263.
Williams, J. G. K., Kubelik, A. R., Livak, K. J., Rafalski, J. A. and Tingey, S. V. (1990). DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucl Acids Res. 18: 6231–6235.
Wolfe, A. D., Xiang, Q. and Kephart, S. R. (1998). Assessing hybridization in natural populations of Penstemon (Scrophulariaceae) using hypervariable intersimple sequence repeat (ISSR) bands. Mol Ecol. 7: 1107–1125.
Zietkiewicz, E., Rafalski, A. and Labuda, D. (1994). Genome fingerprinting by simple sequence repeats (SSR) — anchored polymerase chain reaction amplification. Genomics. 20: 176–183.
Acknowledgements
We wish to thank AgResearch for an NSFT grant, and the University of Canterbury for Research Grant U6223. Further funding came from The Hellaby Grasslands Research Trust, the Hieracium Control Trust and Task Force Green. Suzanne Lambie made the chromosome counts and Flavie Vanlerberghe gave good advice on data analysis. Thanks to Kevin Hogan, Tamsin Braisher and Ashley Sparrow for useful criticisms of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chapman, H., Parh, D. & Oraguzie, N. Genetic structure and colonizing success of a clonal, weedy species, Pilosella officinarum (Asteraceae). Heredity 84, 401–409 (2000). https://doi.org/10.1046/j.1365-2540.2000.00657.x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1046/j.1365-2540.2000.00657.x
Keywords
This article is cited by
-
Genetic diversity, population structure and selection signatures in Enset (Ensete ventricosum, (Welw.) Cheesman), an underutilized and key food security crop in Ethiopia
Genetic Resources and Crop Evolution (2024)
-
Apomixis and genetic background affect distinct traits in Hieracium pilosella L. grown under competition
BMC Biology (2021)
-
Dynamics of apomictic and sexual reproduction during primary succession on a glacier forefield in the Swiss Alps
Scientific Reports (2020)
-
Hybridization rate and genotypic diversity of apomictic hybrids between native (Taraxacum japonicum) and introduced (T. officinale) dandelions in western Japan
Conservation Genetics (2018)
-
Sexual reproduction as a source of ploidy level variation in the model agamic complex of Pilosella bauhini and P. officinarum (Asteraceae: Lactuceae)
Plant Systematics and Evolution (2015)