Abstract
Genetic data from nine species of Merluccius (Euro–African species merluccius, capensis, paradoxus, polli, senegalensis; American species bilinearis, productus, hubbsi, australis) from 21 informative allozyme loci provided insights into the phylogenetic and biogeographical relationships within the genus. The highest values of polymorphic loci and mean heterozygosity occur in the four American species. These values are consistent with large population sizes during speciation (through vicariant processes), and continuing through to the present. Conversely, the lower values of Euro–African species are consistent with bottlenecks occurring during or subsequent to speciation. Euro–African and American species formed two distinct clades. In the former group, merluccius, capensis and senegalensis clustered together as the most derived species, with distinct relationships between polli and paradoxus from an earlier divergence. Similarly, productus, australis and hubbsi clustered closely as the most derived American species, clearly diverging from the more ancestral bilinearis. Analyses including comparative data previously published for M. gayi indicated a close pairing to hubbsi. The data support a north-west Atlantic origin of the genus with unsampled M. albidus of broad Caribbean distribution proposed as the most primitive extant species.
Similar content being viewed by others
Introduction
The genus Merluccius (Rafinesque) (Gadiformes: Merlucciidae) has a broad latitudinal distribution in the Atlantic and Pacific Oceans. In the most complete revision on the genus, Inada (1981) recognized 12 species (Table 1). The existence of an additional species, M. hernandezi from the Gulf of California, has also been indicated but its taxonomic status is uncertain (Cohen et al., 1990). The degrees of divergence, phylogenetic relationships and biogeography within Merluccius have generated considerable speculation (Szidat, 1955; Inada, 1981; Kabata & Ho, 1981).
Two major hypotheses for the origin and dispersal of Merluccius agree that hakes originated in the Atlantic and entered the Pacific through the then-open Panamanian seaway. They differ, however, in details of the origins of the Argentine hake M. hubbsi. A proposed eastern origin of M. hubbsi from a South Pacific stock that rounded Cape Horn to reach Argentina (Szidat, 1955; Inada, 1981) contrasts with a postulated derivation from a western North Atlantic stock (Kabata & Ho, 1981; Ho, 1990).
The morphological similarities among species and the uncertainty of phylogenetic relationships, suggest the application of molecular techniques. These methods have provided critical insights towards resolving similar problems in other teleost taxa. Estimated divergence among fish taxa, traditionally based on morphological measurements, may mask true genetic relationships, which are more precisely reflected by molecular genetic data (Grant, 1987). Allozyme electrophoresis has been a widely applied molecular method for comparing levels of genetic divergence between taxa (e.g. Grant et al., 1999).
Allozyme data have clarified intraspecific relationships in many Merluccius species including M. productus (Utter et al., 1970 and references therein), M. capensis and M. paradoxus (Grant et al., 1988 and references therein), M. merluccius (Roldán et al., 1998 and references therein) and M. hubbsi (Roldán, 1991, 1995). However, only one study, involving three species (Stepien & Rosenblatt, 1996), has examined hakes to investigate species relationships. Our study estimates the level of genetic divergence among 10 Merluccius species and applies this information to infer a molecular phylogeny.
Materials and methods
Sampling
We obtained 2536 specimens representing nine species of Merluccius from America, Europe and Africa (Fig. 1). Specimens were collected directly by factory ships except for the European hake (see details in Roldán et al., 1998) and the Argentinean hake samples, which were taken from research cruises (see details in Roldán, 1995). Samples from two outgroup species, Trisopterus minutus capelanus and Micromesistius potassou (Gadidae), were also collected for the purpose of determining ancestral character states. Species, samples, sample sizes and localities are listed in Table 2.
Electrophoresis
Samples were immediately frozen and were stored at −80°C prior to electrophoretic analyses. Skeletal muscle tissue extraction, electrophoresis, procedures for visualizing proteins and genetic nomenclature generally followed the methods outlined in Roldán et al. (1998). Enzymes surveyed and locus products scored are described in Table 3.
Data analysis
Pairwise multilocus comparisons between samples were calculated by Nei’s genetic distance (1972) and Cavalli-Sforza & Edwards’s (1967) chord distance. A dendrogram was constructed by the unweighted pair-group method with arithmetic averages (UPGMA, Sneath & Sokal, 1973) from the matrix of Nei’s distance. The Fitch and Margoliash algorithm (FM; Fitch & Margoliash, 1967) constructed a tree with unconstrained branch lengths from Cavalli-Sforza and Edwards’s chord distances. The CONTML routine, with the global branch-swapping option produced a maximum likelihood tree based on observed gene frequencies. We evaluated all phylogenetic estimates using a likelihood method with the CONTML program, performing Z-tests (α=0.10) among likelihoods of input-tree topologies to determine if any input tree was statistically more likely than another. Bootstrap values for trees were calculated from 100 replications, resampling across loci. All the above calculations were carried out using the genetic software package PHYLIP version 3.5c (Felsenstein, 1993). A restricted data set was used in some analyses to include data published in Stepien & Rosenblatt (1996) for M. gayi, a tenth species unsampled in this study.
Results
Genetic variability
Among 103 alleles encoded by 23 loci (allele frequencies are available by request from the authors or in the URL entry: http://fc.udg.es/~genetica/LIG/curri/appendix/hake1.html), only two loci (AK* and MDH-2*) were monomorphic for all hake species. Three loci (EST-3*, LDH-B* and LGL*) expressed two alleles, and three or more alleles were detected for the remaining loci. Unbiased gene diversities ranged from 0.030 (SE=0.022) in M. polli to 0.156 (SE=0.045) in M. bilinearis (Table 4).
Genetic divergence among taxa
Conspecific samples grouped together because of the small genetic distances observed (Table 5). In the special case of M. merluccius, Nei’s genetic distances (see details in Roldán et al., 1998) indicate that the degree of divergence between Atlantic and Mediterranean hake does not warrant proposed subspecific distinction (Maurin, 1968; Inada, 1981). Rather, the Mediterranean populations appear to be derived from recent (late or post-Pleistocene) Atlantic colonizations.
For the species of Merluccius Nei’s genetic distance values (Table 5) ranged from 0.122 between M. capensis and M. senegalensis to 1.480 between M. capensis and M. bilinearis. The distance of 0.959 between the two outgroup genera is typical for closely related confamilial genera (Shaklee et al., 1982; Thorpe, 1982). Distances between Miscromesistius or Trisopterus and the species of Merluccius ranged between 1.013 and 2.025. Cavalli-Sforza and Edwards’s chord distances (Table 5) ranged from 0.108 between M. capensis and M. senegalensis to 0.706 between M. capensis and M. bilinearis.
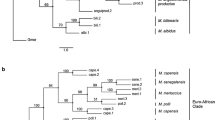
The UPGMA, FM and CONTML trees had very similar topologies and the last two were virtually identical. In all trees, Trisopterus minutus and Micromesistius potassou were placed together as a sister group to the Merluccius species, consistent with Merluccius being a monophyletic genus. All trees placed M. australis, M. hubbsi, M productus and M. bilinearis (American species) in one cluster and M. capensis, M. merluccius, M. paradoxus, M. polli and M. senegalensis (Euro–African species) in another. Merluccius capensis and M. senegalensis are sister taxa and are grouped with European hake M. merluccius (Fig. 1).
Based on maximum likelihood evaluations of tree topologies, the CONTML topology was significantly more likely than any other (ln L=UPGMA: 285.110; FM: 413.057; CONTML: 504.704). The confidence limits of the CONTML tree identified the same areas of instability indicated by the bootstrapped values for the UPGMA tree where the node connecting M. hubbsi to M. australis and M. productus cluster collapsed. Two other nodes collapsed, one connecting M. polli to M. paradoxus and the other connecting M. paradoxus to the outgroup.
Discussion
Genetic variability within species
Patterns of within-species genetic variability (Table 4) yield some initial insight into ancestral relationships and demographic events during and following divergence. The sensitivity of alleles per locus to sample sizes coupled with the large variability of sample sizes among species may partially explain the absence of a clear pattern of alleles per locus.
Notably, the highest values of polymorphic loci and mean heterozygosity, parameters more robust to variable sample sizes (Nei, 1975), occur in the four American species. These values are consistent with large population sizes during speciation (through vicariant processes), and continue through to the present (Grant et al., 1988; Futuyma, 1998). Conversely, the lower percentages of polymorphic loci and mean heterozygosities of Euro–African species are consistent with bottlenecks occurring during or subsequent to speciation.
Phylogenetic hypotheses and biogeography
Our study’s limitation to nine of the 12 presently recognized species of Merluccius restricts full phylogenetic consideration of the genus. However, the relationships indicated by the CONTML tree provide some clear phylogenetic guidelines. The clustering and branching order of the Euro–African hakes were clearly resolved. Merluccius polli and M. paradoxus was the most divergent lineage and M. capensis, M. senegalensis and M. merluccius the most derived lineage.
Within the American group, the position of M. bilinearis was distinct and it can be considered the most ancient species. We sought further clarification in this group through analysing data from the study of Stepien & Rosenblatt (1996), which included data for M. gayi. Using M. hubbsi and M. productus common to both studies as reference species, compatible data from 16 allozyme loci were assembled based on similar tissue expression and allele frequencies. The grouping of M. gayi with other American species (Fig. 1) clearly relates it to its geographical neighbours. Its pairing with M. hubbsi is consistent with the pairing of M. australis and M. productus indicated by our data (Table 5).
A biochemical clock calibrated for hakes may provide an approximate time frame for species separation. Under an allozyme clock developed from fish studies, Nei’s genetic distance D=1.0 represents ≈19 Myr of separation, as calibrated empirically by genetic distances between taxa separated by the rise of the Panama Isthmus (Vawter et al., 1980) or between transoceanic taxa founded after the opening of the Bering Strait (Grant, 1987). Under this calibration, average values of D=1.411 between Trisopterus and Merluccius species and D=1.644 between Micromesistius and Merluccius species correspond to a time since divergence of 26–31 million years BP in the Oligocene epoch. This interval contains the collision of Africa with Eurasia, followed by the Tethys Sea remaining open to shallow-water fishes until about 20 Myr ago.
This chronology agrees with fossil records. A considerably larger number of gadoid genera appeared in the Miocene deposits of central Europe and the Caucasus than in the Oligocene (Svetovidov, 1948), confirming the presence of a common ancestor of both families in the Tethys Sea. If this large-scale geological event influenced Merlucciidae–Gadidae divergence, then we would expect to find similar effects in other groups of fishes. For example, the geographical distributions and divergence between the sardine genera Sardina and Sardinops and lophiid genera Lophius and Lophiomus are also compatible with this explanation (Parrish et al., 1989; Grant & Leslie, 1993).
Merlucciidae is the only family of supragadoids with continuous latitudinal distribution, and its presence in the New Zealand area (and that of the gadoid Micromesistius australis) has been explained as a fragmented and formerly continuous Gondwanan distribution of the ancestral merluciid lineage (Howes, 1991 and references therein). The degree of genetic divergence between New Zealand and Patagonian populations of M. australis is unknown. Nevertheless, we expect some differentiation as a result of restricted gene flow. Significantly lower frequencies of the GPI-1*125 allele among New Zealand populations are apparent from data reported by Smith et al. (1979). Presumably, gene flow is in one direction from the New Zealand area to Patagonia given past and present flow patterns between these regions. The most likely time for migration is during the extended pelagic juvenile phase, when young fish are at the mercy of prevailing currents. During this time, the flow of the strong Antarctic Circumpolar Current leads to the north, and a significant part of the current branches northward flowing up the west coast of South America. Alternatively, regional gyre-like circulations may reduce or prevent such circumpolar migration, as proposed for larval retention within shelf and bank populations of Atlantic cod (Ruzzante et al., 1998).
The average D=0.980 between American species and Euro–African species corresponds to a time since divergence of 18 Myr BP in the Miocene epoch. Merluccius-like fossils have been found from the Upper Oligocene to the Lower Miocene in central European deposits (M. lednevi and M. errans) (Svetovidov, 1948). Geological events that cause fragmentation of the contiguous, ancestral distribution are considered to be the major means of distributional pattern formation (Nelson & Platnick, 1981). We suggest that the first split in the ancestral Atlantic population of Merluccius followed the separation of the South American and African Plates (Fig. 2). Merluccius polli and M. paradoxus in the eastern Atlantic represent the descendants of an early Old World Merluccius clade.
For the two southern African hakes, M. capensis and M. paradoxus, Grant et al. (1988) reported Nei’s (1972) genetic distance of c0.583 (SE=0.160), representing can estimated time interval from 7.6 to 13.6 Myr ago. Our data (D=0.754; 14.3 Myr BP) are consistent with a divergence time at the upper range of this interval. Grant et al. (1988) suggested two biogeographical scenarios that may explain the sympatric distribution of these fishes in southern Africa: (i) past episodes of oceanic cooling displaced northward and isolated ancestral populations of southern African Merluccius; and (ii) the two species represent different biogeographical dispersals of North Atlantic taxa along the west coast of Africa. Our data (Fig. 1) support the second hypothesis, indicating a genetic affinity between M. capensis, and M. senegalensis and M. merluccius, two species with northern distributions. Accordingly, M. capensis appears to be the more recent of the two southern African hakes, derived jointly with M. senegalensis from a recent divergence event different from that leading to the presently sympatric M. paradoxus. A more ancient divergence best explains the relationships of M. paradoxus and M. polli.
Conclusions
In summary, our data provide further insight into the uncertain current understanding of phylogeny and biogeography of Merluccius. With regard to Euro–African species, our data support distinct divergences of presently sympatric South African species, as noted above. The recent derivation of M. merluccius indicated by our data agrees with a similar conclusion by Ho (1990). These findings support the origin of the genus beyond the present eastern North Atlantic (Ho, 1974, 1990) but contrast with hypotheses of this region being the original source of the genus (Inada, 1981; Kabata & Ho, 1981).
The uniformly higher within-species variation of American hake (Table 4) supports consistently large biomasses throughout their generic evolutionary histories. The contrasting lower values of Euro–African species, particularly the more-anciently diverged M. paradoxus and M. polli, are consistent with dispersive origins from an American source including bottleneck events during and after speciation.
Within the American species, M. bilinearis in the western North Atlantic is considered the most ancient species with an estimated divergence time of 12 Myr BP in the Miocene. We propose that M. gayi, M. productus and M. australis arose by a dispersal route along the Pacific American coast before the closure of the Panama Isthmus occurred 3.5 Myr BP (Fig. 2). This possibility is supported by M. productus fossils being common in the Pliocene deposits of California (Fitch, 1969). The pairing of M. gayi and M. hubbsi (Fig. 1) is consistent with a dispersal hypothesis for their origins (Szidat, 1955; Inada, 1981) contrasted with a proposed wide separation of these species (Ho, 1990). But the possibility exists that M. hubbsi reached its present distribution by a simultaneous dispersal route along the Atlantic coast, as proposed by Ho (1990).
The absence of molecular data from two American species restricts further phylogenetic and biogeographical details being proposed for American species. Given the pairing of M. productus and M. australis we anticipate a similar genetic proximity of unsampled M. angustimanus with an overlapping range of M. productus. Similarly, the broad western Atlantic distribution of unsampled M. albidus suggests the possibility of this species being the most divergent and ancient Merluccius taxon, consistent with Ho (1990). For the moment, the present data provide new insights and suggest further possibilities that must be tested by collection of additional data.
References
Cavalli-Sforza, L. L. and Edwards, A. W. F. (1967). Phylogenetic analysis: models and estimation procedures. Evolution, 32: 550–570.
Cohen, D. M., Inada, T., Iwamoto, T. and Scialabba, N. (1990). FAO Species Catalogue vol. 10, Gadiform Fishes of the World (Order Gadiformes). An Annotated and Illustrated Catalogue of Cods, Hakes, Grenadiers and Other Gadiform Fishes Known to Date. FAO Fisheries Synopsis no. 125, vol. 10. FAO, Rome.
Felsenstein, J. (1993). PHYLIP (Phylogeny inference package), version 3.5c. Distributed by the author, Department of Genetics, University of Washington, Seattle.
Fitch, J. E. (1969). Fossil records of certain schooling fishes of the California Current system. Calif Coop Oceanic Fish Invest Report, 13: 71–80.
Fitch, W. M. and Margoliash, E. (1967). Construction of phylogenetic trees. Science, 155: 279–284.
Futuyma, D. J. (1998). Evolutionary Biology, 3rd edn. Sinauer Associates, Sunderland, MA.
Grant, W. S. (1987). Genetic divergence between congeneric Atlantic and Pacific Ocean fishes In: Ryman, N. and Utter, F. M. (eds). Population Genetics and Fishery Management, pp. 225–246 University of Washington Press, Seattle.
Grant, W. S. and Leslie, R. W. (1993). Biochemical divergence and biogeography of anglerfish of the genus Lophius (Lophiiformes). J Zool, 231: 465–485.
Grant, W. S., Becker, I. I. and Leslie, R. W. (1988). Evolutionary divergence between sympatric species of southern African Hakes, Merluccius capensis and M. paradoxus I. Electrophoretic analysis of proteins. Heredity, 61: 13–20.
Grant, W. S., García-Marín, J. L. and Utter, F. M. (1999). Defining population boundaries for fishery management In: Saleem, M. (ed.). Genetics in Sustainable Fisheries Management, pp. 27–71 Blackwell Science, Oxford.
Ho, J. -S. (1974). Parasitic copepods as indicator of hake’s (Merluccius) zoogeography. Proc 3rd Int Cong Parasitol, 3: 1633–1634.
Ho, J. -S. (1990). Phylogeny and biogeography of hakes (Merluccius; Teleostei): a cladistic analysis. U S Fish Bull, 88: 95–104.
Howes, G. J. (1991). Biogeography of gadoid fishes. J Biogeog, 18: 595–622.
Inada, T. (1981). Studies on the Merlucciid fish. Bull Far Seas Fish Res Laboratory, Shimizu, Japan, 18: 1–172.
Kabata, Z. and Ho, J. -S. (1981). The origin and dispersal of hake (genus Merluccius: Pisces: Teleostei) as indicated by its copepod parasites. Oceanogr Mar Biol Ann Rev, 19: 381–404.
Maurin, C. (1968). Les merlus des côtes nord et nord-ouest d’Afrique (Atlantique et Méditerranée) Ph.D. Thesis, Université de Nancy, France.
Nei, M. (1972). Genetic distance between populations. Am Nat, 106: 283–292.
Nei, M. (1975). Molecular Population Genetics and Evolution, Elsevier, New York.
Nei, M. (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics, 41: 225–233.
Nelson, G. and Platnick, N. (1981). Systematics and Biogeography, Cladistics and Vicariance. Columbia University Press, New York.
Parrish, R. H., Serra, R. and Grant, S. W. (1989). The monotypic sardines, Sardina and Sardinops: their taxonomy, distribution, stock structure and zoogeography. Can J Fish Aquat Sci, 46: 2019–2036.
Roldán, M. I. (1991). Enzymatic polymorphisms in the Argentinian hake, Merluccius hubbsi Marini, of the Argentinian continental shelf. J Fish Biol, 39 (Suppl. A), 53–59.
Roldán, M. I. (1995). Relaciones filogenéticas en el género Merluccius y estructura genética poblacional en la merluza argentina (Merluccius hubbsi) y la merluza europea (M. merluccius). Ph.D. Thesis, University of Girona, Spain.
Roldán, M. I., García-Marín, J. L., Utter, F. M. and Pla, C. (1998). Population genetic structure of European hake, Merluccius merluccius. Heredity, 81: 327–334.
Ruzzante, D., Taggart, C. T. and Cook, D. (1998). A nuclear DNA basis for shelf- and bank-scale population structure in northwest Atlantic cod (Gadus morhua): Labrador to George Bank. Mol Ecol, 7: 1663–1681.
Shaklee, J. B., Tamaru, C. S. and Waples, R. S. (1982). Speciation and evolution of marine fishes studied by the electrophoretic analysis of proteins. Pacific Sci, 36: 141–157.
Smith, P. J., Patchell, G. J. and Benson, P. G. (1979). Glucosephosphate isomerase and isocitrate dehydrogenase polymorphisms in the hake, Merluccius australis. N Z J Mar Freshw Res, 13: 545–547.
Sneath, P. H. A. and Sokal, R. R. (1973). Numerical Taxonomy. W. H. Freeman, San Francisco, CA.
Stepien, C. and Rosenblatt, R. H. (1996). Genetic divergence in antitropical pelagic marine fishes (Trachurus, Merluccius and Scomber) between North and South America. Copeia, 3: 586–598.
Svetovidov, A. N. (1948). Treskoobraznye (Gadiformes). Fauna SSSR Zool Institut Akademi Nauk SSSR 34, Ryby (Fishes), 9(4): 1–222 (In Russian; English translation, 1962, Israel Program for Scientific Translations, Jerusalem).
Szidat, L. (1955). La fauna de parásitos de Merluccius hubbsi como carácter auxiliar para la solución de los problemas sistemáticos y zoogeográficos del género Merluccius L. Comun Inst Nac Invest Cienc Nat, B Aires, 3: 1–54.
Thorpe, J. (1982). The molecular clock hypothesis: Biochemical evolution, genetic differentiation and systematics. Ann Rev Ecol Syst, 13: 139–168.
Utter, F. M., Stormont, C. and Hodgins, H. (1970). Esterase polymorphism in vitreous fluid of Pacific hake, Merluccius productus. Anim Blood Groups biochem Genet, 1: 69–82.
Vawter, A. T., Rosenblatt, R. H. and Gorman, G. C. (1980). Genetic divergence among fishes of the Eastern Pacific and the Caribbean: support for the molecular clock. Evolution, 34: 705–711.
Acknowledgements
We thank Peter Beerli, University of Washington for helpful discussions on data analysis techniques and Ju-Shey Ho for useful comments on the manuscript. M.I.R. was supported by a postgraduate fellowship from the Agencia Española de Cooperación Internacional. This work was partially supported by agreement 160719 with Instituto Español de Oceanografía.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Roldán, M., García-Marín, J., Utter, F. et al. Genetic relationships among Merluccius species. Heredity 83, 79–86 (1999). https://doi.org/10.1038/sj.hdy.6885300
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6885300
Keywords
This article is cited by
-
Phylogenetic prospecting for cryptic species of the genus Merluccius (Actinopterygii: Merlucciidae)
Scientific Reports (2021)
-
Clarifying the taxonomic status of Merluccius spp. in the northeastern Pacific: a combined morphological and molecular approach
Reviews in Fish Biology and Fisheries (2011)
-
Molecular Organization and Evolution of 5S rDNA in the Genus Merluccius and Their Phylogenetic Implications
Journal of Molecular Evolution (2009)