Abstract
Co-operative behaviour may evolve by enhancing the genetic similarity of group members. Increased group similarity is thought to be the basis for the ‘subsocial route’ of social evolution in the spider family Eresidae. Two processes may promote the similarity of individuals within populations or breeding groups, namely philopatry in stable environments and founder events in a stochastic environment. We show that both processes led to genetic differentiation within and among populations of the subsocial spider Stegodyphus lineatus. Within populations we distinguished between the genetic structure caused by random mating and philopatry in old breeding groups and that caused by newly founded groups consisting of sibs. Such sib-groups suggest that new breeding groups are established primarily by single females. The different gene coancestries among breeding groups resulted in high variances among single-locus data. The results imply that sex-specific dispersal behaviour (random male mating-dispersal or female group founding) had different impacts on the population structure. This type of population structure, where within-population philopatry and founder events may lead to differential proliferation of breeding groups, is very similar to that presumed for social spiders, and is also one that could provide the conditions for interdemic selection.
Similar content being viewed by others
Introduction
Breeding tactics and social structure influence the population genetic structure of species. In particular, social living can lead to structured populations, with a large degree of genetic differentiation between populations or social groups. It is less clear to what extent genetic structure may facilitate the evolution of social behaviour (Van Staaden, 1995). Some models of the evolution of social behaviour stress the importance of increasing the genetic identity of group members (e.g. Hamilton, 1964a,b; Wade & Breden, 1981; Michod, 1993). However, increasing genetic similarity may lead to inbreeding depression, and thus lower individual fitness. Inbreeding may also reduce the effect of within-group similarity relative to the population at large, owing to an overall decrease in genetic variability (Michod, 1993). The costs of inbreeding may be balanced or outweighed by the cost of migration. High costs of migration can favour philopatry, i.e. the tendency to stay in a home area, and facilitate both genetic differentiation and sociality. Alternatively, founder events, and especially population founding by propagules of related females, may have profound effects on the direction of social evolution by increasing genetic variance among populations. Founder events and high population turnover may lead to the fixation of new behaviours and to speciation (Templeton, 1980).
In spiders, high population turnover and inbreeding are associated with the occurrence of co-operative behaviour (Avilés, 1997). The social spiders exhibit an extreme form of population structure. They live in closed breeding colonies and new colonies are formed either by fission of an existing colony, by swarming of related females, or by single gravid females (Lubin & Robinson, 1982; Vollrath, 1982). Unlike nonsocial species, males do not appear to disperse large distances. Gene flow between established colonies is thought to be rare: genetic analyses have found fixation of alternative alleles in neighbouring colonies (Lubin & Crozier, 1985; Roeloffs & Riechert, 1988; Smith & Engel, 1994; Smith & Hagen, 1996). Given a high probability of colony extinction, frequent colony turnover may enhance intercolony selection. It seems likely that behavioural preadaptations leading to tolerance and co-operation among colony members, on the one hand, and population structure on the other, would promote conditions suitable for maintaining sociality. Which of these conditions preceded sociality remains unknown.
Several spider species are considered to be subsocial: they experience both maternal brood care beyond the first few developmental instars that is typical of most spider species and an extended phase of tolerance among young within the maternal nest (Buskirk, 1981). However, they live solitarily as adults and do not live in closed colonies. Sociality in the spider family Eresidae is thought to have evolved via the ‘subsocial pathway‘ by a prolongation of an early tolerance phase without dispersal. The subsocial route to sociality entails increasing the genetic variance among breeding groups by means of philopatry or a restricted breeding system. Johannesen et al. (1998) investigated the genetic structure of the ground-dwelling subsocial eresid Eresus cinnaberinus (Eresidae) and found that it had a genetic structure intermediate to those of social and asocial spiders. Limited dispersal and vertical maternal lineages created high genetic variance between neighbourhoods within populations.
Stegodyphus lineatus is a subsocial eresid, whose life history differs substantially from that of E. cinnaberinus. Stegodyphus lineatus is an annual organism. It builds its nests and webs in bushes and trees, and is often found in unstable environments such as dry watercourses (wadis) that may be periodically flooded (Ward & Lubin, 1993). In contrast, E. cinnaberinus lives underground for three to four years in silk-lined burrows. Males and females mature at different ages, which largely prevents sib–sib mating. The habitat of E. cinnaberinus generally experiences little annual variation and populations are known to persist for many years (Baumann, 1997).
The aim of this study was to investigate the genetic and breeding structure of populations of S. lineatus in order to determine the degree of genetic differentiation within and among populations. In addition to addressing the breeding and genetic structure of a species showing preadaptations for social behaviour, the study provided an opportunity to compare S. lineatus and E. cinnaberinus, two subsocial species in the same family but with different life histories. We asked how are populations of S. lineatus structured genetically? (i) Is there evidence of dispersal of males and females and does philopatry of either sex lead to within-population differentiation? (ii) What is the extent of local mating and do males mate outside the natal group? (iii) Do founder events play a role in enhancing genetic variance among groups or populations?
Ecological investigations of dispersing young of S. lineatus have shown initial dispersal distances of only 2 m (Lubin et al., 1998). Only 32% of large juveniles and subadults moved their nest site, with a median distance of 2.24 m (Ward & Lubin, 1993). It is unclear how far adults disperse upon reaching sexual maturity and whether they avoid mating with close relatives. It is also unclear to what extent dispersal distances are related to food availability. Ward & Lubin (1993) found increased dispersal under good food conditions, but Schneider (1995) showed in the laboratory that food availability can sustain tolerance. Nonrandom dispersal may be influenced by preferential selection of shrubs with dense branch architecture (Ward & Lubin, 1993). In both S. lineatus and E. cinnaberinus males leave their nests and seek mates during a short breeding season (Schneider & Lubin, 1996; Baumann, 1997).
In the present study we investigated the distribution of S. lineatus adults following juvenile dispersal. We seek to distinguish between female and male dispersal. Males may influence gene flow significantly among established groups, but only females can found new populations or groups.
Materials and methods
Collections
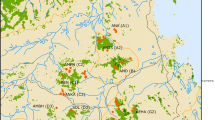
Spiders were collected from six patches in the Judean and Negev deserts in Israel in May 1997 (Fig. 1). The average distance between patches was 40 km. One site, Revivim, consisted of two clusters of acacia trees (Acacia raddiana) separated by ≈500 m of desert (patches RA and RB). At both Revivim and Beer Sheva, the spiders were aggregated in trees (Fig. 2). These groups of spiders were used for analyses of relatedness among spiders within and among the trees and for the analysis of within-patch genetic structure. The remaining three patches, Avdat, Upper Wadi Zin, and Arad were included as outgroup populations in an overall population genetic analysis. Here, spider nests were mostly situated singly in low shrubs in the wadis or on the nearby slopes.
The Beer Sheva patch was scheduled for a housing development, and we sampled the entire population. At Revivim we collected half of each tree population, and 25% of the estimated total population. Arad and Avdat wadis were searched intensively and all encountered spiders were collected. Therefore, although sample sizes of these populations vary, they represent a total estimate of genetic variation. The Upper Zin Wadi population, which is being used for behavioural studies, consisted of about 400 individuals. From the entire wadi we obtained a random sample of 23 males.
Electrophoretic analysis
Spiders were taken live to the laboratory and stored at –80°C. We screened a total of 43 enzyme systems. Twenty-one of these systems representing 25 loci could be scored consistently: Aat-1, -2 (EC 2.6.1.1), Adh (EC 1.1.1.1), Ak (EC 2.7.4.3), Apk (EC 2.7.3.3), Est (EC 3.1.1.-), Fum (EC 4.2.1.2), G-3-Pdh (EC 1.2.1.12), G-6-Pdh (EC 1.1.1.49), Gpd (EC 1.1.1.8), Hk (EC 2.7.1.1), Idh (EC 1.1.1.42), Ldh-1, -2 (EC 1.1.1.27), Mdh (EC 1.1.1.37), Pep-A (glycine–leucine) and Pep-B-1, -2 (leucine–glycine–glycine) (EC 3.4.11 or 13), Pgd (EC 1.1.1.44), Pgi (EC 5.3.1.9), Pgm (EC 5.4.2.2), Pk (EC 2.7.1.40), Sorbdh-1, -2 (EC 1.1.1.14), Tpi (EC 5.3.1.1).
Electrophoretic analysis was performed by cellulose acetate electrophoresis (Hebert & Beaton, 1993). Four buffer systems were used: Tris-Maleic acid pH=7.8 (Richardson et al., 1986) for Aat, Gpd, Mdh, Pgi, Pgm; Tris-Maleic acid pH=7.0 (adjusted with maleic acid to pH=7.0 from TM pH=7.8) for Pk, Sorbdh, Tpi, G-6-Pdh, Ldh, Est; Tris-Citrate pH=8.2 (Richardson et al., 1986) for Ak, Apk, Idh, Fum, Est, G-3-Pdh, Hk, Pgd; and Tris-Glycine pH=8.5 (Hebert & Beaton, 1993) for Adh, Pep-A, Pep-B.
All enzymes were run for 30 min at 250 V, except Aat, Ldh, Est (TM pH=7.0), G-3-Pdh, Apk, Pgi, Pgm, which were run for 45 min. During the initial screening, esterase was run in three different buffers, and during analysis in two, to check for compound effects on banding patterns. Three loci were detected, of which only one showed consistent banding patterns in all buffers. Only this single monomeric esterase locus was included in the analysis.
All spider tissue samples were homogenized in 80 mL Pgm-buffer (Harris & Hopkinson, 1978) by ultrasound and centrifuged at 15800 g for 2 min. Posterior abdominal tissue was used for all enzymes except Aat, Ak, Pgd and Pk for which leg and thorax tissue were used.
Genetic analyses
Putative loci were analysed only if they displayed patterns consistent with known quaternary structure (Richardson et al., 1986). A locus displaying only one band was interpreted as monomorphic. Alleles were designated according to their relative mobility (most common allele = 1.00). Loci were tested for departure from expected Hardy–Weinberg proportions with the Louis & Dempster (1987) exact test. All patches, and the tree populations (spider groups) within the Beer Sheva and Revivim patches, were tested for genotypic linkage disequilibria with the program GENEPOP (Raymond & Rousset, 1995). Genetic differentiation within and among spider groups and patch populations was analysed with the estimates of FIS, FIT and FST by employing the F-statistics of Weir & Cockerham (1984). Standard deviations for these estimates were obtained by jackknifing over the polymorphic loci. A hierarchical distribution of genetic variation was quantified with Wright's (1978) hierarchical F-statistics. Isolation by distance (Slatkin, 1993) was investigated among all six patches, and among spider groups within the Beer Sheva and Revivim populations. Isolation by distance within and among patches was tested by correlating pair-wise logarithmic GST values (Nei, 1977) with log-geographical distance, and the significance of pair-wise estimates was tested with a Mantel test (1000 permutations, MANTEL subroutine in GENEPOP). The genetic relationship within and among populations was estimated by the maximum likelihood method with the program CONTML (PHYLIP, Felsenstein, 1993). This method analyses the distribution of allele frequencies and assumes that branch lengths are determined by drift alone.
To test the hypothesis that males disperse more than females we used two approaches. First, we tested for sex-differential F-statistics. If FIT(males) < FIT(females) this would indicate that males move more than females. We tested this prediction using a two-tailed t-test. Secondly, if the males found in a group consist of foreign and local group males, we may expect males within a group to be less related to each other than females are to one another. We tested for relatedness differences by comparing the average group-wide relatedness, R, of males and females, respectively, and by analysing the relatedness difference of males and females within a single group. The former relatedness prediction was tested using a two-tailed t-test. The latter relatedness prediction assumes an estimate significantly greater than zero, and was tested by a one-tailed t-test. Relatedness estimates of spider group members were investigated using the program RELATEDNESS 4.2c (Queller & Goodnight, 1989). We tested the population genetic structure for both males and females at Beer Sheva. Too few males were available to test the Revivim population.
One of our aims was to investigate the population genetic structure in S. lineatus for evidence of philopatry or group founding behaviour. If either occurs, spiders may cluster in breeding groups. Because standard F-statistics at the within-population level are biased by the breeding structure (Chesser, 1991a), we analysed populations and individuals at Beer Sheva and Revivim under different sampling assumptions of clustering, and of sib and nonsib-groups. Relatedness of group individuals may either be caused by family relationships or by the Wahlund effect. In diploid organisms, if mating within a group is random the group inbreeding estimate Fg=(He – Ho)/He ≈ 0, where He is the expected and Ho the observed group heterozygosity. However, if a group consists largely of sibs it will always have an excess of heterozygotes, and Fg (comparable to FIT for a single population, or FIS for multiple groups) will always be negative (Rasmussen, 1979 [NB: positive in Rasmussen's approach]). To distinguish between within-patch genetic structure caused by drift–inbreeding (Wahlund effect) and that caused by sib-groups we compared Fg estimates within single spider groups, to the relatedness, R, of the same group relative to the population estimate of FIT. Taken together, R(group) > 0 and Fg < 0 indicate relatedness of group members because of sampling largely sibs, despite FIT > 0; whereas R(group) > 0, Fg=0 and FIT > 0 together imply relatedness of group members because of drift effects caused by population substructure. Fg-values were tested for deviation from random association of alleles, Fg=0, by 100 permutation tests over alleles using the program GENETIX (Belkhir et al., 1996–98). This test can be compared to a bootstrap over alleles.
Results
We found relatively few males in the populations sampled (Table 1), probably through sampling late in the reproductive season (males mature in March–April; Schneider, 1997).
The amount of genetic variation was within the range typical for arthropods, He ≈ 0.10 (averaged over 24 loci). A total of 11 loci were polymorphic at the 95% level in at least one population (Aat-1, -2 (each 3 alleles), Adh (2), Ak (2), Est-1 (4), Fum (2), Idh (3), Ldh-2 (3), Pep-A (3), Pep-B1 (4), Sorbdh-2 (2) (allele frequency tables provided on request); He ≈ 0.25 over 11 loci, Table 1. Despite large differences in sample sizes there were no differences in expected heterozygosity among samples. The Avdat population had fewer alleles per locus than the remaining five samples. There was no indication of genotypic linkage disequilibria in any population and all loci polymorphic at the 95% level were included in analyses of genetic differentiation.
Relatedness of group individuals
The average relatedness of spider group members in the Beer Sheva population was R=0.22 (Table 1). However, FIT > 0 indicated a population substructure (Table 2). At first sight, these findings suggest that the relatedness of group members was caused by drift effects in a substructured population. However, if the group R=0.50, which indicates full-sibs, and group Fg is significantly lower than zero, then relatedness could be explained by sib-groups. Two likely sib-groups were found, BS3 (R=0.54, Fg=−0.51) and BS7 (R=0.51, Fg=−0.42) (Table 1). Based on multilocus comparisons, the seven BS3 individuals probably consisted of six full-sibs, and either one foreign or half-sib individual. BS7 probably consisted of three sibs. If BS3 and 7 are removed from the analysis, the population R declines to 0.15, and the Beer Sheva population FIT increases to 0.12. This indicates that R > 0 for the remaining groups was caused by drift–inbreeding. Thus, different family backgrounds of the groups confounded the population estimate at Beer Sheva.
Relatedness estimates for Revivim A and B were not significantly different from zero. One group, RB4 (R=0.23, Fg=−0.13) may have consisted of some sibs. Otherwise the data suggest that relatedness of group individuals was caused by philopatry and that group members originated from several parent couples.
No significant differences in the relatedness of females and males were found (Table 3). Males of a group were on average more related to each other than were females (R(males)=0.34 and R(females)=0.28). However, the population inbreeding estimate FIT was significantly lower in males than in females (P< 0.01) indicating that females are more restricted in their dispersal than males. However, high variances of male F-statistics parameters suggest that the groups sampled were of different genetic background. If some of the males we sampled were nondispersed kin, and others were not, this would explain the observed negative FIS estimates, and corroborate R (males) > 0.
Within-population genetic structure
We calculated Hardy–Weinberg proportions including all polymorphic loci and for loci under the expectation of at least one rare homozygote. Deviation from expected Hardy–Weinberg proportions was found in the Beer Sheva population with both approaches (P< 0.05). The Revivim patches (A + B) did not deviate significantly from Hardy–Weinberg proportions. However, when we only included the five loci where a rare homozygote is expected, the significance increased from P=0.50 to P=0.10. No significant deviations were found in the remaining three populations using exact tests, but when applying a permutation test, the Arad population showed a significant excess of heterozygotes (Fg=−0.18; P< 0.05) (Table 1).
The within-population genetic structure was investigated for the spider groups. At Beer Sheva, high single-locus variances caused the F-statistic estimates to be nonsignificant. However, as shown above, the high variances were caused by unequal sampling of spider groups consisting of sibs (BS3 and 7), and of spider groups consisting of several family groups. Removing the two sib-groups led to FIT > 0, thus corroborating the Hardy–Weinberg estimates for the Beer Sheva population. Although not significantly different from zero, the positive FIS values now indicate sampling several sib-groups per tree. In both Revivim patches FIS ≈ 0 implied that individuals from the spider groups, on average, originated from several parent-pairs. Correcting for differences in group membership at Beer Sheva, the amount of subdivision within Revivim (A + B), and Beer Sheva was similar, FIT ≈ 0.12.
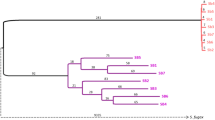
At both Beer Sheva and Revivim we sampled from trees that stood singly and in three cases, trees whose branches intertwined (Fig. 2). In all three cases groups from intertwining trees clustered (Fig. 3). However, in one case, RB2 and 3, the branch length was not significantly greater than zero. At Revivim groups from the two patches (A and B) clustered within their respective patches.
Maximum likelihood phenograms of relationships among groups of Stegodyphus lineatus living in trees within Beer Sheva, Revivim A and B, and among all samples including the remaining populations, Arad, Avdat and Upper Zin Wadi. *Branch lengths significantly different from zero within Beer Sheva and Revivim (95% confidence intervals).
Isolation by distance among groups was observed at Beer Sheva (P< 0.01) and Revivim (A + B) (P< 0.001). The result for Beer Sheva may arise because of a bias caused by groups BS5–7 (see cluster analysis above). When these three groups were pooled and the Beer Sheva population was reanalysed, a near significant (P=0.06) isolation by distance was found. Removing the two sib-group clusters, BS3 and 7, did not alter the isolation by distance result (P< 0.05). The result at Revivim is a nonequilibrium consequence of sampling from two independent population units, RA and RB.
Genetic structure among populations
The genetic differentiation among populations (N=6 populations, pooled data) was about equal to the differentiation calculated among all samples (N=19, groups): FST(pooled)=0.12 and FST(groups)=0.14 (Table 2). Two hierarchical analyses of genetic variance (Wright, 1978) were performed (Table 2). The first estimate was calculated including only the spider groups from Beer Sheva and Revivim, and showed that 2/3 of the variance was distributed among groups within either Beer Sheva or Revivim. The second estimate, now including Upper Wadi Zin, Avdat and Arad, showed that half the variance was distributed within patches relative to the total population (all samples). The lower within-patch estimate of genetic variance (0.106–0.073) in the latter analysis may be explained in part by the fact that Upper Wadi Zin, Avdat and Arad were single samples and no within-patch variance was estimated.
No isolation by distance was found among the six populations (P > 0.30). The three outgroup populations, Arad, Avdat and Upper Wadi Zin, seemed to cluster randomly within the spider groups at Beer Sheva and Revivim (Fig. 2).
Discussion
Population units
Increased identity of group and population members was obtained both by isolation among populations, and by philopatry within populations. Among populations, FST defined for populations (FST(pooled)) and for all groups (FST(groups)) showed near identical values of FST ≈ 0.12–0.14. This indicates that the different patches constitute population units, and that the spider groups were not population units. If the spider groups had constituted separate population units, one would expect FST (pooled) < FST (groups), because drift among spider groups would homogenize allele frequencies in the pooled data set. Wright's hierarchical FST analysis showed that the amount of variance found among populations decreased considerably when including the three outlier populations (Upper Wadi Zin, Arad and Avdat). Also, groups from a single population tended to cluster together, and the variance among groups within a population (Beer Sheva or Revivim) was lower than the variance among populations. These results indicate that each population of spider groups has primarily a single origin (with possible later immigration), and that populations constitute independent units. Thus, for S. lineatus, patches, not spider groups, defined populations. This contrasts with the social spiders where colonies or clusters of colonies, and not patches, most often define the population units (Smith & Engel, 1994; Smith & Hagen, 1996; but see also Lubin & Crozier, 1985).
No isolation by distance was found among populations of S. lineatus. Isolation by distance assumes an equilibrium between genetic drift and gene flow, and requires a constant habitat relative to dispersal distance (Slatkin, 1993). The lack of isolation by distance implies a high turnover and founder events of S. lineatus populations, and suggests that greater gene flow occurs within populations and among wadis than is apparent. Clustered sampling locations (Upper Wadi Zin and Avdat; RA and RB; see Fig. 1) may at least have led to a nonequilibrium isolation by distance. Ecological data indicate that extreme annual fluctuations indeed are common in this species and local extinction may be a common phenomenon (Y. Lubin, pers. obs.). Within populations, philopatry has created population subdivision. Given the combination of founder events and within-population philopatry occurring in a fluctuating environment, some groups of related individuals may proliferate while others may go extinct, and thereby enhance the genetic variance among mating-groups. This demographic structuring is very similar to that presumed for social spiders, and is also one that could provide the conditions for interdemic selection (Avilés, 1993,1997).
Breeding and within-population structure
The within-population substructure of S. lineatus can probably be explained by two processes, namely an interaction between philopatry and group founding. Evidence for the philopatric behaviour of S. lineatus was suggested by FIT > 0, and a within-patch isolation by distance of spider groups (see also Fig. 3). Group founding is suggested by the two groups BS3 and 7, which showed an extreme heterozygote excess. Gene flow between groups no doubt takes place, but group founding and philopatry within groups must predominate in creating a viscous population structure leading to differentiation among groups.
We may get an idea of how the population structure at Beer Sheva has evolved by considering the two groups BS3 and 7, which consisted of sibs, suggesting that (single) females dispersed to adjacent breeding sites. Stegodyphus lineatus is an annual organism and the adult spiders of the groups we collected were about to reproduce. The siblings would form a breeding group of individuals with common ancestors. This scenario suggests that females of S. lineatus occasionally disperse and that genetic differentiation within populations is caused by female dispersal followed by random mating, predominantly within the newly founded isolate. Random mating within isolates of philopatric sibs will enhance the differentiation among groups in the following generation. If this scenario is correct we must distinguish between male mating-dispersal and females founding new populations. Data from the present study on male genetic structure are inconclusive because of high variances and small sample sizes. On the one hand, significantly lower differentiation estimates suggest higher dispersal than for females, but the high variances and relatedness estimates, on the other hand, suggest that males were sampled from groups of different genetic background, maybe because of the sampling of both dispersed and nondispersed individuals.
Sampling spider groups of unequal genetic background (sib and random mating groups) led to high single-locus variance estimates among the groups, and this may lead to a statistically nonsignificant structuring estimate. Traditional F-statistics based on the island model assume random mating and noncorrelated gene coancestries of populations or breeding groups (Chesser, 1991b). In the first generation of a newly founded isolate, Fg < 0 because the sibs are defined as a random mating group (Rasmussen, 1979; Chesser, 1991a). In the next generation, despite random inbreeding among sibs (given no immigration), Fg ≈ 0, because the individuals are as related as they are inbred (Cockerham, 1973). Thus, the negative Fg = −0.18 at Arad, where we sampled the entire population of a small wadi, may indicate that a high proportion of the individuals were sibs. Previously, negative FIS estimates have been found in newly formed (first generation) groups of the beetle Phalacrus substriatus, after dispersal of related gravid females (Ingvarsson, 1998). If inbreeding is pervasive but mating is random among group members, as is likely to be the case in the social spiders, the average subpopulation inbreeding estimate FIS will not indicate inbreeding because all individuals within a breeding colony, on average, should be equally heterozygotic. FIT was shown to be an appropriate inbreeding estimator in social spiders (Smith & Engel, 1994; Smith & Hagen, 1996) for inferring genetic structure among colonies within a sample site. In this context, we define inbreeding as increased relatedness of group members relative to a panmictic index; the patch population. Genetic drift in substructured populations will decrease the inbreeding effective population size but increase its variance effective population size (Chesser, 1991b). Taken to the extreme, as in social spiders, where the population (colony) turnover is high, even the variance effective population size may decrease as a result of losing all variation through intercolony selection. In contrast, dispersal and outbreeding in S. lineatus maintains genetic variation.
Similar to S. lineatus, high levels of genetic variation, high genetic differentiation of populations and increased genetic identity of individuals within neighbourhoods was also found in the subsocial eresid spider E. cinnaberinus. However, the cause of population subdivision in the latter species seems to be fundamentally different. In a transect sample of E. cinnaberinus (Johannesen et al., 1998), clusters of nests within a larger neighbourhood were comprised largely of sib-groups. This caused the FIS estimate among clusters to become negative, but, in contrast to the Beer Sheva and Revivim populations of S. lineatus, no patch substructure was observed relative to mating probability; thus FIT=0. This was achieved primarily because the sexes of the same clutch do not mature in the same year. However, a subdivision estimate among neighbourhoods on the same hill was obtained because of site tenacity of females, giving FST > 0, whereas FIT and FIS were slightly (but not significantly) negative (Johannesen et al., 1998). If breeding groups are known and are defined as populations the average inbreeding estimate will be slightly negative (Cockerham, 1969). The extremely limited female dispersal in E. cinnaberinus led to high genetic differentiation between populations on different hills.
Thus, there seem to be differences in dispersal and breeding systems between the two subsocial species. If group founding by females primarily determines within-patch structure in S. lineatus, and if male mating-dispersal preferentially takes place in the vicinity of the natal site, then this species may exhibit a form of female-mediated population structure approaching that found in the social spiders.
References
Avilés, L. (1993). Interdemic selection and sex ratio: a social spider perspective. Am Nat, 142: 320–345.
Avilés, L. (1997). Causes and consequences of cooperation and permanent-sociality in spiders. In: Choe, J. and Crespi, B. (eds) Social Competition and Cooperation Among Insects and Arachnids, II Evolution of Sociality, pp. 476–498. Cambridge University Press, Cambridge.
Baumann, T. (1997). Populationsökologische und zönotische Untersuchungen zur Bedeutung von Habitatqualität und Habitatfragmentierung für Spinnenpopulationen auf Trockenrasen am Beispiel von Eresus cinnaberinus (Oliv. 1789). Ph.D. Dissertation, Universität Bremen, Germany.
Belkhir, K., Borsa, P., Goudet, J., Chikhi, L. and Bonhomme, F. (1996–98). GENETIX logiciel sous WindowsTM pour la génétique des populations Laboratoire Génome et Populations, CNRS UPR 9060, Université de Montpellier, France.
Buskirk, R. (1981). Sociality in the Arachnida. In: Hermann, H. R. (ed.) Social Insects, pp. 282–367. Academic Press, New York.
Chesser, R. K. (1991a). Gene diversity and female philopatry. Genetics, 127: 437–447.
Chesser, R. K. (1991b). Influence of gene flow and breeding tactics on gene diversity within populations. Genetics, 129: 573–583.
Cockerham, C. C. (1969). Variance of gene flow frequencies. Evolution, 23: 72–84.
Cockerham, C. C. (1973). Analyses of gene frequencies. Genetics, 74: 679–700.
Felsenstein, J. (1993). PHYLIP (Phylogeny Inference Package), Version 3.5c. Department of Genetics, University of Washington, Seattle.
Hamilton, W. D. (1964a). The genetical evolution of social behavior. I. J Theor Biol, 7: 1–16.
Hamilton, W. D. (1964b). The genetical evolution of social behavior. II. J Theor Biol, 7: 17–52.
Harris, H. and Hopkinson, D. A. (1978). Handbook of Enzyme Electrophoresis in Human Genetics. North-Holland Publishing Company, Amsterdam.
Hebert, P. D. N. and Beaton, M. J. (1993). Methodologies for Allozyme Analysis using Cellulose Acetate Electrophoresis. Helena Laboratories, Beaumont, TX.
Ingvarsson, P. K. (1998). Kin-structured colonization in Phalacrus substriatus. Heredity, 80: 456–463.
Johannesen, J., Baumann, T., Seitz, A. and Veith, M. (1998). The significance of relatedness and gene flow on population genetic structure in the subsocial spider Eresus cinnaberinus (Araneae: Eresidae). Biol J Linn Soc, 63: 81–98.
Louis, E. J. and Dempster, E. R. (1987). An exact test for Hardy–Weinberg and multiple alleles. Biometrics, 43: 805–811.
Lubin, Y. D. and Crozier, R. H. (1985). Electrophoretic evidence for population differentiation in a social spider Achaearanea wau (Theridiidae). Insect Soc, 32: 297–304.
Lubin, Y. D. and Robinson, M. H. (1982). Dispersal by swarming in a social spider. Science, 216: 319–321.
Lubin, Y. D., Hennicke, J. and Schneider, J. (1998). Settling decisions of dispersing Stegodyphus lineatus (Eresidae) young. Isr J Zool, 44: 217–225.
Michod, R. E. (1993). Inbreeding and evolution of social behavior. In: Thornhill, N. W. (ed.) The Natural History of Inbreeding and Outbreeding: Theoretical and Empirical Perspectives pp. 74–96. University of Chicago Press, Chicago.
Nei, M. (1977). F-statistics and analysis of gene diversity in subdivided populations. Ann Hum Genet, 41: 225–233.
Queller, D. C. and Goodnight, K. F. (1989). Estimating relatedness using genetic markers. Evolution, 43: 258–275.
Rasmussen, D. I. (1979). Sibling clusters and genotypic frequencies. Am Nat, 113: 948–951.
Raymond, M. and Rousset, F. (1995). GENEPOP (v. 1.2): population genetics software for exact tests and ecumenicism. J Hered, 86: 248–249.
Richardson, B. J., Baverstock, P. R. and Adams, M. (1986). Allozyme Electrophoresis. A Handbook for Animal Systematics and Population Studies. Academic Press, San Diego, CA.
Roeloffs, R. and Riechert, S. E. (1988). Dispersal and population-genetic structure of the cooperative spider, Agelena consociata in West African rainforest. Evolution, 42: 173–183.
Schneider, J. (1995). Survival and growth in groups of a subsocial spider (Stegodyphus lineatus). Insect Soc, 42: 237–248.
Schneider, J. M. (1997). Timing of maturation and the mating system of the spider, Stegodyphus lineatus (Eresidae): how important is body size? Biol J Linn Soc, 60: 517–525.
Schneider, J. M. and Lubin, Y. (1996). Infanticidal male eresid spiders. Nature, 381: 655–656.
Slatkin, M. (1993). Isolation by distance in equilibrium and non-equilibrium populations. Evolution, 47: 264–279.
Smith, D. R. and Engel, M. S. (1994). Population structure in an Indian cooperative spider, Stegodyphus sarasinorum Karsch (Eresidae). J Arachnol, 22: 108–113.
Smith, D. R. and Hagen, R. H. (1996). Population structure and interdemic selection in the cooperative spider Anelosimus eximius. J Evol Biol, 9: 589–608.
Templeton, A. R. (1980). The theory of speciation via the founder principle. Genetics, 94: 1011–1038.
van Staaden, M. J. (1995). Breeding tactics, social structure and genetic variation in mammals: problems and prospects. Acta Theriologia, Suppl3: 165–182.
Vollrath, F. (1982). Colony formation in a social spider. Z Tierpsychol, 60: 313–324.
Wade, M. J. and Breden, F. (1981). Effect of inbreeding on the evolution of altruistic behavior by kin selection. Evolution, 35: 844–858.
Ward, D. and Lubin, Y. (1993). Habitat selection and the life history of a desert spider, Stegodyphus lineatus (Eresidae). J Anim Ecol, 62: 353–363.
Weir, B. S. and Cockerham, C. C. (1984). Estimating F-statistics for the analysis of population structure. Evolution, 38: 1358–1370.
Wright, S. (1978). Evolution and the Genetics of Populations, vol. 4, Variability Within and Among Natural Populations University of Chicago Press, Chicago.
Acknowledgements
We thank Ofer Eifan, Tamar Erez and Ori Segev for helping with field collections. Volker Loeschcke, Jutta Schneider and Michael Veith made valuable comments on earlier versions of the manuscript. This work was supported by a DFG-Graduiertenkolleg stipend to Jes Johannesen. This is publication no. 270 of the Mitrani Department of Desert Ecology.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Johannesen, J., Lubin, Y. Group founding and breeding structure in the subsocial spider Stegodyphus lineatus (Eresidae). Heredity 82, 677–686 (1999). https://doi.org/10.1046/j.1365-2540.1999.00528.x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1046/j.1365-2540.1999.00528.x
Keywords
This article is cited by
-
Dispersal behaviour and colony structure in a colonial spider
Behavioral Ecology and Sociobiology (2012)
-
Cooperative prey capture by young subsocial spiders: II. Behavioral mechanism
Behavioral Ecology and Sociobiology (2005)