Abstract
As part of a programme to integrate the genetic and physical chromosome maps of Brassica oleracea we have carried out fluorescence in situ hybridization (FISH) to mitotic and meiotic chromosomes of B. oleracea var. alboglabra, using repeat sequence DNA probes. We have confirmed that there are three 18S-5.8S-25S rDNA sites in the haploid genome and accurately determined their locations for the first time; two occur subtelomerically on the short arms of the two satellited acrocentric chromosomes (chromosomes 4 and 7), whereas the third site is adjacent to the centromere on the short arm of a large submetacentric chromosome (chromosome 2). 5S rDNA sequences are located on the long arm of this same chromosome, with closely adjacent major and minor loci. A highly repeated sequence (pBcKB4) colocalizes with the pericentromeric heterochromatin on all chromosomes, but six chromosomes exhibit very strong signals whereas the remaining three show only weak signals. The construction of a partial karyotype for B. oleracea var. alboglabra, based on several major cytogenetical landmarks provided by FISH localization of repetitive DNA sequences, provides a valuable framework for the future physical mapping of nonrepetitive sequences.
Similar content being viewed by others
Introduction
One of the goals of investigating the genomes of important crop species such as Brassica oleracea is the integration of genetic maps constructed by RFLPs and PCR-based markers with corresponding physical maps produced by karyotype analysis and cytogenetical determination of the positions of DNA sequences on chromosomes.
The chromosome number of B. oleracea (2n=18) was established by Morinaga (1934). Further analysis by U (1935) established the relationship between B. oleracea, B. campestris and B. nigra and their amphidiploids, and is widely known as U's triangle. Pachytene analysis of the three basic diploid species was carried out by Röbbelen (1960) who described pachytene chromosome structure and characterized the karyotypes on the basis of structural characteristics of total chromosome length, symmetry of the arms and the shape of the heterochromatic pericentromere regions. He found that the largest chromosome of B. oleracea was twice the size of the smallest with the remaining chromosomes showing a gradation of lengths. He classified them into one very long chromosome, five long chromosomes, two medium chromosomes and one short chromosome. The arm ratios classified the haploid karyotype into six metacentric chromosomes, two submetacentric and one ‘subterminal’ (acrocentric) chromosome with a satellite, which was described as the nucleolar chromosome. Olin-Fatih & Heneen (1992) described the karyotype of B. oleracea var.acephala cv. Hammenhögs extra krusbladiga, and grouped it into four ‘median’ (metacentric), four ‘submedian’ (submetacentric) and one ‘subterminal’ (acrocentric) pair. Like Röbbelen, they identified a single satellited nucleolus organizing chromosome and placed it in the submetacentric group. Cheng et al (1995) published a revised karyotype for B. alboglabra including three ‘median’, four ‘submedian’ and two ‘subterminal’ chromosomes. This revision excluded the satellite of the nucleolar chromosome in the calculation of the centromeric index and thus altered the classification of this nucleolar chromosome from the submedian group to the subterminal group
Investigations of the mitotic karyotype, produced from root-tip preparations, are limited by the small size of the Brassica chromosomes indicated by the haploid 1C genome size of 0.8 pg (Bennett & Leitch, 1995), which makes it difficult to identify chromosomes especially following metaphase accumulation by spindle inhibitors. Preparations of Brassica chromosomes from anthers have a number of advantages over root-tip preparations. First, synchronous mitotic divisions of tapetal cells can produce good quality elongated metaphase and prometaphase chromosomes which have not been blocked by spindle inhibitors. Secondly, meiotic prophase I chromosomes from pollen mother cells (PMCs) are much more extended than mitotic chromosomes and also have specific and clear chromomere patterns. Thus they should provide better material for the development of a physical map based on in situ hybridization of molecular markers (Shen et al., 1987; Albini & Schwarzacher, 1992; Xu & Earle, 1996). Finally, other stages of meiosis, for example diakinesis, can provide useful information about chiasma distribution and frequency, which can be used to derive genetic distances for marked chromosomes.
The development of fluorescent in situ hybridization (FISH) has meant that it is possible to locate DNA sequences directly on chromosomes to produce physical chromosome maps. Earlier studies have established the numbers, but not the positions of loci, for 18S-5.8S-25S (45S) and 5S rDNA, and also analysed a repetitive DNA present in pericentromeric heterochromatin of Brassica mitotic chromosomes (Maluszynska & Heslop-Harrison, 1993; Harrison & Heslop-Harrison, 1995; Brandes & Heslop-Harrison, 1996). In this paper we determine, for the first time, the chromosomal positions of 45S rDNA and 5S rDNA sequences for Brassica oleracea var. alboglabra, which figures importantly as a parent in one of the current B. oleracea genetic mapping populations.
Materials and methods
Plant materials
Brassica oleracea var.alboglabra line DHdA12 is a genetically homozygous doubled-haploid line derived from B. oleracea var.alboglabra by microspore culture (Bohoun et al., 1996). This line has the advantages of a short generation time and self-compatibility. Also, it is a relatively erect and robust plant unlike other short-cycle Brassicas. Line DHdA12 is a parent in a series of genetical mapping exercises (including Ramsay et al., 1996) which is a further justification for the choice of this line for molecular cytogenetic analysis. The plants were grown under normal conditions in a glasshouse.
Fixation
Flower buds were taken from plants and carefully dissected under a stereo-microscope. Single anthers from individual buds in the size range 0.5–2 mm were removed and the meiotic stages of single anthers checked by lacto-proprionic orcein squashing. The remaining anthers were fixed in Carnoy's fixative (6 parts ethanol: 3 parts chloroform: 1 part glacial acetic acid) at room temperature and stored at 4°C for up to 6 months.
Preparation of slides
The method of Zhong-Xiao-Bo et al. (1996) for spreading PMCs was modified for Brassica material as follows: anthers were removed from Carnoy's fixative and placed in 3:1 fixative (ethanol: glacial acetic acid) for 10 min. This fixative was replaced twice more, in order to remove all traces of chloroform, which affects the efficiency of the enzymes in the next stage. The anthers were washed three times for 5 min in citrate buffer (10 mM, pH 4.5) and then digested in enzyme mixture 0.3% (w/v) cellulase, 0.3% (w/v) pectinase and 0.3% cytohelicase (w/v) (Sigma) in 10 mM citrate buffer pH 4.5 for 2–3 h in a moist chamber at 37°C. Enzyme digestion was stopped by replacing the enzyme solution with cold distilled water. The anthers were then placed on ice. A single anther was placed on a glass slide and 20 μL of 60% acetic acid added. The anther was broken up with a fine needle and the slide placed on a hot block (45°C) for 1 min. After this time the slide was placed on a bench and the material fixed by adding cold 3:1 ethanol acetic fix (200 μL) initially in a circle around the acetic acid drop, until the fixative and acetic acid were thoroughly mixed. The slide was tilted, flushed with fresh fixative and then air-dried and stored with desiccant at −20°C for up to 6 months.
Probes
The following DNA clones were used in FISH experiments.
(I) Clone pTa71 (Gerlach & Bedbrock, 1979) containing a 9 kb EcoRI fragment of Triticum aestivum L. consisting of the 18S-5.8S-25S rRNA genes and the nontranscribed intergenic spacer regions.
(II) Plasmid pCT 4.2 containing a 500 bp 5S rDNA insert cloned from Arabidopsis thaliana (ABRO USA).
(III) Plasmids pBoKB1 derived from B. oleracea and pBcKB4 derived from B. campestris containing repetitive DNA clones from the pericentromeric heterochromatin (Harrison & Heslop-Harrison, 1995).
Plasmids I and II were grown and DNA extracted by the method of Sambrook et al. (1989). Inserts from pBoKB1 and pBcKB4 were amplified by PCR using Pharmacia M13 primers. The DNA sequences pCT 4.2 and pTa71 were labelled with biotin-dUTP or FITC dUTP by nick translation using the protocol of the manufacturer (Boehringer Mannheim). The probes pBoBK1 and pBcBK4 were labelled by a second PCR incorporating biotin-dUTP.
In situ hybridization
Fluorescence in situ hybridization was carried out according to the method described by Fransz et al. (1996), with the following additions: before the paraformaldehyde fixation step, slides were incubated in 0.01 g pepsin in 100 mL of 0.01 M HCl for 10 min at 37°C and then washed three times in 2×SSC for 5 min
Between 50 and 100 ng of probe were used for each slide, made up to 20 μL in hybridization fluid containing 50% deionized formamide, 2×SSC and 10% dextran sulphate.
The probe mixture was applied to the slide and sealed under a 22×22 mm coverslip using vulcanizing solution. The slide was placed on a hot block at 75°C for 4 min, which denatured the target DNA and probe simultaneously, and hybridized overnight at 37°C in a damp chamber.
Posthybridization washes were performed as described by Fransz et al. (1996). Detection of biotin-labelled probes by Texas Red antiavidin and amplification by biotin-conjugated goat antibody was according to manufacturer's instructions (Vector). FITC-labelled probes were amplified by a FITC detection system according to manufacturer's instructions (Cambio).
Slides were counterstained with DAPI (1 μg/mL) in Vectashield antifade mounting medium (Vector).
Photomicroscopy
Slides were examined by means of a Nikon Eclipse T300 microscope. Capture and analysis of images was achieved using an image analysis system (Vysis Ltd).
Karyotype analysis
Twenty good-quality mitotic metaphases, probed with either 45S or 45S and 5S rDNA and counterstained with DAPI were selected for measurements of the karyotype. The captured metaphases were measured directly on the screen. The chromosome relative lengths (% of haploid complement) and centromeric indices (short arm/total chromosome length×100) were calculated for each metaphase. The lengths of the satellites were excluded following the practice in karyotype construction in other species such as Allium (de Vries et al., 1990)
Results
FISH
All the probes used in this study were successfully applied both to meiotic stages and mitotic chromosomes from B. oleracea anthers.
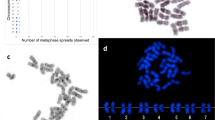
Clone pTa71 (45S rDNA) hybridized to three loci on three different mitotic metaphase chromosomes. Two of these loci are near the short-arm ends of the two acrocentric chromosomes in the complement (nominally 4 and 7), coinciding with the positions of secondary constrictions in DAPI-stained preparations. In var. alboglabra, there is an obvious difference in the copy number of the 45S rDNA between chromosome 4 (small signal) and chromosome 7 (large signal). The third locus is situated in the middle of the short arm of a large submetacentric chromosome (nominally chromosome 2) with a signal size similar to that on chromosome 4 (Fig. 1a,b). In diakinesis/metaphase I stages the three hybridization signals can be seen associated with three different bivalents (Fig. 1).
In situ hybridization to chromosomes of Brassica oleracea var. alboglabra. (a) Tapetal mitotic metaphase stained with DAPI, showing three pairs of loci for 45S rDNA detected by FITC. (b) Tapetal mitotic metaphase, showing three pairs of loci for 45S rDNA detected by Texas Red and a single 5S pair of rDNA signals detected by FITC. (c) Meiotic pachytene. 45S rDNA loci detected by Texas Red. Two paired loci are coalesced and a third paired locus forms a single signal. Major (large arrow) and minor (small arrow) 5S rDNA paired loci detected by FITC. (d) Meiotic diakinesis showing nine bivalents. Distribution of pBcKB4 detected by Texas Red. Six pairs of chromosomes are strongly labelled and three pairs of chromosomes are only weakly labelled. Three paired loci of 45S rDNA labelled with FITC. (e) Tapetal mitotic metaphase showing the distribution of pBcKB4 detected by Texas Red. Three pairs of loci of 45S rDNA detected by FITC. Scale Bar=10 μm.
5S rDNA is located just distal to the centromere in the long arm of the largest submetacentric chromosome (nominally chromosome 2) (Fig. 1b). Although only a single locus is generally observed in mitotic preparations, in pachytene preparations the chromosomes are much longer and 5S rDNA can be seen to have two closely adjacent major and minor loci (Fig. 1c). In pachytene spreads clumping of the pericentromeric heterochromatin (DAPI bright regions) can be seen to form a synizetic knot.
pBoKB1 and pBcKB4 gave signals at sites which colocalized with the DAPI bright pericentromeric heterochromatin. The pBoKB1 signals were relatively weak (not shown) compared to pBcKB4 which gave clear signals on six chromosome pairs, whereas three pairs labelled only very weakly and can only be detected using a single bypass filter for Texas Red in mitotic spreads and at diakinesis (Fig. 1d,e). The signals covered the entire pericentromeric regions.
Karyotype
A partial karyotype of B. oleracea var . alboglabra (Fig. 2), was constructed based on measurements of 20 good-quality tapetal mitotic metaphases (Table 1), which had been hybridized with 45S rDNA alone or both 45S and 5S rDNA and counterstained with DAPI (Fig. 1a,b). The largest chromosome is approximately twice the length of the smallest, whereas the remainder show a gradation of lengths in between these extremes. The combination of length rank position, centromere location and repetitive DNA markers allows several of the chromosomes to be identified with some confidence from DAPI-stained preparations hybridized with the 45S probe. Three chromosomes, can be positively identified by virtue of their 45S rDNA loci and their modal rank positions are 2, 4 and 7. Chromosomes 4 and 7 can be further distinguished by the size of the rDNA signal. However, it is clear that these marked chromosomes do not always occupy these rank positions; they are merely the modal rank positions. This illustrates the need for caution in attempting to identify the other, unmarked, chromosomes. Chromosome 1 can be identified with some confidence because chromosome 2, which carries a 45S rDNA cluster, hardly ever (1/20) ranked as the largest chromosome and in addition is distinctly submetacentric (mean CI 30.93±3.59). Rank position chromosome 3 lies in between two 45S rDNA-marked chromosomes (2 and 4), which therefore gives some confidence in identifying this chromosome. However, some chromosomes (e.g. 5 vs. 6; 8 vs. 9) are sufficiently similar to have a significant risk of misclassification through ‘reversal of order’ (Matérn & Simak, 1968). The development of appropriate FISH probes should eliminate these uncertainties, which is of course one of the principal aims of this study.
Pachytene chromosomes were found to be particularly useful in that they are very extended compared to mitotic chromosomes and they reveal clear chromomere patterns. On the other hand, there is a tendency for a synizetic knot to form at this stage which involves clumping of the pericentromeric heterochromatin and thus prevents resolution of individual chromosomes. However, well-spread pachytene preparations gave good resolution of distal chromosome regions protruding from the pachytene clumps.
Discussion
Anther preparations were shown to provide a number of advantages for chromosome analysis in B. oleracea compared to conventional root-tip preparations. In addition to pachytene stage PMCs with extended bivalents, they produce large quantities of mitotic chromosomes of good morphology from tapetal cells. In the B. oleracea var.alboglabra plants that we examined, tapetal cell divisions appear to be most frequent at zygotene, and disappear rapidly as pachytene progresses.
Following our procedure of excluding the satellites from the measurements, we found that there were seven metacentric/submetacentric pairs and two pairs of satellited acrocentric chromosomes. There is little correspondence between the chromosome numbering that we propose and that put forward by Olin-Fatih & Heneen (1992) and Cheng et al. (1995), because our numbering is based on ranked lengths only, whereas they grouped the chromosomes according to centromere position.
Pachytene chromosomes provide good chromomere patterns, particularly at telomeric regions, which should be particularly useful to unite chromosome maps and genetic maps in this species. Such an approach has been demonstrated for meiotic chromosomes in the tomato (Xu & Earle, 1995, 1996). In addition, extended prophase I chromosomes give a higher resolution of closely adjacent sites which can be detected by FISH. The two adjacent 5S rDNA loci can be more clearly resolved in pachytene chromosome preparations. The use of pachytene preparations in order to resolve DNA target sites for loci that are close together in a chromosomal subregion has previously been reported for human loci (Bello et al., 1989, Hultén & Armstrong, 1994). FISH provides an efficient method for identifying the number and physical location of repeated sequences. We have confirmed the presence of three loci for 45S rDNA in B. oleracea previously described by Maluszynska & Heslop-Harrison (1993) and Snowdon et al. (1997) and described by these authors as two ‘major’ and one, barely detectable, ‘minor’ sites. Snowdon et al. failed to find a third locus in metaphases but showed a small signal in interphase cells. Although we observed differences in the sizes of these loci in our cultivar, not even the smallest signal could be described as a ‘minor’ site. More importantly, we have determined the chromosomal positions of these loci. Two occur subtelomerically in the short arms of the acrocentric chromosomes and the third locus has been placed in the short arm of the largest submetacentric chromosome, close to the centromere.
In most organisms, the bulk of the 5S rDNA is organized into tandem arrays found at a small number of chromosomal locations (Long & Dawid, 1980). We observed two closely adjacent 5S rDNA sites in B. oleracea var. alboglabra, located near to the centromere in the long arm of the largest submetacentric chromosome. Although two loci have been reported for this gene by Brandes & Heslop-Harrison (1996), they did not determine their positions, and so this is the first report of 5S rDNA location for B. oleracea.
The probe for a pericentromeric heterochromatin-associated repeated DNA sequence isolated from B. oleracea was found to give only a weak FISH signal on our alboglabra material. On the other hand, the probe for a pericentromeric heterochromatin-associated repeat derived from B. campestris gave very clear signals on six chromosomes and weaker signals on three chromosomes. This distribution is slightly different from that observed by Harrison & Heslop-Harrison (1995), who found seven pairs of sites in B. oleracea with the probe pBcKB4. The variation observed with regard to 45S rDNA loci and the pericentromeric repeat probe may be associated with the cultivar B. oleracea var.alboglabra examined by us in comparison to the cultivar Savoy cabbage Sabanda (Acc. no. OB420) (Maluszynska & Heslop-Harrison, 1993) and B. oleracea cv. Brauncsch-Weiger (Snowdon et al., 1997) for 45S rDNA and cultivar Dwarf green curled for pBcKB4 (Harrison & Heslop-Harrison, 1995).
In conclusion, it is clear from the results that we have obtained to date, that FISH will prove an ideal tool with which to integrate the cytological and genetical maps of B. oleracea. The karyotyping exercise reported in this paper, together with the localization by FISH of repeated DNA sequences, has resulted in a basic framework for further physical mapping (Fig. 2). Currently, we are in the process of developing the technology required to exploit low copy number and unique sequences in order to integrate fully genetic maps (e.g. Howell et al., 1996; Ramsay et al., 1996) with the physical chromosome map. This is of particular importance in the case of Brassica species, where a plethora of independent numbering systems has developed for both chromosomes and linkage groups. We will adopt the nomenclatural framework of Sharpe et al. (1995) and Parkin et al. (1995), which provides a standard set of designations for both the linkage groups of B. napus (N1-N19) and those of the component B. rapa and B. oleracea genomes.
References
Albini, S. M. and Schwarzacher, T. (1992). In situ localization of two repetitive DNA sequences to surface-spread pachytene chromosomes of rye. Genome, 35: 551–559.
Bello, M. J., Salagnon, N., Rey, J. A., Guichaoua, M. A., Bergelefranc, J. L., Jordan, B. R. et al (1989). Precise in-situ localisation of NCAM ET51 and D11529 on human meiotic chromosomes. Cytogenet Cell Genet, 52: 7–10.
Bennett, M. D. and Leitch, I. J. (1995). Nuclear DNA amounts in Angiosperms. Ann Bot, 76: 113–176.
Bohoun, E. J. R., Keith, D. J., Parkin, I. A. P., Sharpe, A. G. and Lydiate, D. J. (1996). Alignment of the conserved C-genomes of Brassica oleracea and Brassica napus. Theor Appl Genet, 93: 833–839.
Brandes, A. and Heslop-Harrison, J. S. (1996). Comparative analysis of the large scale organisation of Crucifer genomes. BBSRC Warwick Meeting. Plant and Animal Genome Analysis Abstracts, 10.
Cheng, B. F., Heneen, W. K. and Chen, B. Y. (1995). Mitotic karyotypes of Brassica campestris and Brassica alboglabra and identification of the B. alboglabra in an addition line. Genome, 38: 313–319.
de Vries, J. N. (1990). Onion chromosome nomenclature and homeology relationships — Workshop Report. Euphytica, 49: 1–3.
Fransz, P. F., Maike, S., Montijn, B., Tenhoopen, R., Wiegant, J., Kooter, J. M. et al (1996). Detection of single-copy genes and chromosome rearrangements in Petunia hybrida by fluorescence in situ hybridization. Plant J, 9: 767–774.
Gerlach, W. L. and Bedbrock, J. R. (1979). Cloning and characterisation of ribosomal RNA genes from wheat and barley. Nucl Acids Res, 7: 1869–1885.
Harrison, G. E. and Heslop-Harrison, J. S. (1995). Centromeric repetitive DNA sequences in the genus Brassica. Theor Appl Genet, 90: 157–165.
Howell, P. M., Marshall, D. F. and Lydiate, D. J. (1996). Towards developing intervarietal substitution lines in Brassica napus using marker assisted selection. Genome, 39: 348–358.
Hultén, M. A. and Armstrong, S. J. (1994). Oocyte mapping of chromosome 9. Third International Workshop on Chromosome 9. Ann Hum Genet, 58: 217
Long, G. O. and Dawid, I. B. (1980). Repeated genes in eukaryotes. Ann Rev Biochem, 49: 727–774.
Maluszynska, J. and Heslop-Harrison, J. S. (1993). Physical mapping of rDNA loci in Brassica species. Genome, 36: 774–781.
Matérn, B. and Simak, M. (1968). Statistical problems in karyotype analysis. Hereditas, 59: 280–288.
Morinaga, T. (1934). Interspecific hybridisation in Brassica. VI. The cytology of F1hybrids of B. juncea and B. nigra. Cytologia, 6: 62–67.
Olin-Fatih, M. and Heneen, W. G. (1992). C-banded karyotypes of Brassica campestris, B. oleracea, and B. napus. Genome, 35: 283–589.
Parkin, I. A. P., Sharpe, A. G., Keith, D. J. and Lydiate, D. J. (1995). Identification of the A and C genomes of amphidiploid Brassica napus (oilseed rape). Genome, 38: 1122–1131.
Ramsay, L. D., Jennings, D. E., Bohoun, E. J. R., Arthur, A. E., Lydiate, D. J., Kearsey, M. J. et al (1996). The construction of a substitution library of recombinant backcross lines in Brassica oleracea for the precision mapping of quantitative trait loci. Genome, 39: 558–567.
Röbbelen, G. (1960). Beiträge zur Analyse des Brassica-Genoms. Chromosoma, 11: 205–228.
Sambrook, J., Fritsch, E. F. and Maniatis, T. (1989) Molecular Cloning — A Laboratory Manual Cold Spring Harbor Laboratory Press, NY.
Sharpe, A. G., Parkin, I. A. P., Keith, D. J. and Lydiate, D. J. (1995). Frequent nonreciprocal translocations in the amphidiploid genome of oilseed rape (Brassica napus). Genome, 38: 1112–1121.
Shen, D., Wang, Z. and Wu, M. (1987). Gene mapping on maize pachytene chromosomes by in situ hybridization. Chromosoma, 95: 311–314.
Snowdon, R. J., Köhler, W. and Köhler, A. (1997). Chromosomal localization and characterization of rDNA in the Brassica A and C genomes. Genome, 40: 582–587.
U, N. (1935). Genome analysis of Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilisation. Jap J Bot, 7: 389–452.
Xu, J. and Earle, E. D. (1995). Direct FISH of 5S rDNA on tomato pachytene chromosomes places the gene at the heterochromatic knob immediately adjacent to the centromere of chromosome 1. Genome, 39: 216–221.
Xu, J. and Earle, E. D. (1996). High resolution physical mapping of 45S (5.8S, 18S and 25S) rDNA gene loci in the tomato genome using a combination of karyotyping and FISH of pachytene chromosomes. Chromosoma, 104: 545–550.
Zhong-Xiao-Bo, Z., Dejong, H. J. Zabel, P. (1996). Preparation of tomato meiotic pachytene and mitotic chromosomes suitable for fluorescence in situ hybridisation (FISH). Chromosome Res, 4: 24–28.
Acknowledgements
We acknowledge the gift of the probes pBoKB1 and pBcKB4 from Professor J. S. Heslop-Harrison, John Innes Centre, Norwich, UK. This work is supported by BBSRC grant GO4051.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Armstrong, S., Fransz, P., Marshall, D. et al. Physical mapping of DNA repetitive sequences to mitotic and meiotic chromosomes of Brassica oleracea var. alboglabra by fluorescence in situ hybridization. Heredity 81, 666–673 (1998). https://doi.org/10.1046/j.1365-2540.1998.00432.x
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1046/j.1365-2540.1998.00432.x
Keywords
This article is cited by
-
FISH painting for chromosome identification of aneuploid cauliflower (Brassica oleracea L. var. botrytis)
The Nucleus (2023)
-
Fluorescent labelling of in situ hybridisation probes through the copper-catalysed azide-alkyne cycloaddition reaction
Chromosome Research (2016)
-
Assigning Brassica microsatellite markers to the nine C-genome chromosomes using Brassica rapa var. trilocularis–B. oleracea var. alboglabra monosomic alien addition lines
Theoretical and Applied Genetics (2012)
-
Fluorescence in situ hybridization on plant extended chromatin DNA fibers for single-copy and repetitive DNA sequences
Plant Cell Reports (2011)
-
Modified protocol for obtaining isolated and high-resolution pachytene chromosomes
The Nucleus (2011)