Abstract
Field elm (Ulmus minor) is a riparian tree that grows in rare, small populations scattered along temporary watercourses in the Balearic Islands, nowadays mostly covered with Mediterranean vegetation. Agriculture and farming on the fertile land along the periodically flooded plains have reduced the elm populations to sparse tree lines along the creek beds. The presence of field elm in this very anthropic landscape has led some authors to consider it as an introduced species in the Balearics. However, pollen data suggest these elms may be the remains of larger populations experiencing continuous population shrinkage during the Holocene, and hence be native to the isles. In this paper, we apply genetic markers to assess whether field elm is or is not indigenous to the Balearic Islands. We compare the genetic variation in nine nuclear microsatellites of six Balearic populations (three in each of the largest islands, Majorca and Minorca) with that of three natural Iberian populations located in two regions, one geologically (Baetic mountains, SE Iberia) and another historically (Catalonia, NE Iberia) related to the islands. Principal coordinates analysis and Bayesian clustering methods reveal a strong genetic differentiation of the Balearic populations from the Iberian ones, and even among islands, which support their native origin. Genotypic variation in the islands is very low and clonal reproduction is very high compared with the mainland, as it is frequently observed in populations of clonal species where sexual reproduction is limited. We discuss the practical implications of these findings for the conservation of elm genetic resources of these findings.
Similar content being viewed by others
Introduction
Conservation commonly focuses on protection of the interspecific diversity of a region. A key concept underpinning this, is that of ‘nativeness’, namely whether a given species has a natural distribution in a given place. Species that are native to an area usually have a higher conservation value afforded to them, than species that have been introduced by man. The distinction is blurred in cases were non-native species provide some ecological or societal benefit, and particularly where species have been deliberately introduced outside of their native range for conservation purposes. Nevertheless, the typical situation is that native species are commonly included in conservation planning and introduced species are not. In many cases, there is little controversy about whether a species is native or introduced. However, in some cases there is uncertainty and this creates uncertainty for conservation planning.
Webb (1985) noted that ‘too often it would appear that the assignment [of native state] has been made either by copying uncritically from earlier works or on essentially intuitive grounds’. To address this problem, Webb (1985) made a systematic attempt to establish a set of criteria for reducing the subjectivity in assessing whether a species is native or introduced. These criteria included historical, ecological and genetic features; specifically, fossil evidence of former existence, historical evidence of introduction, possible means of introduction, occurrence in natural versus man-made habitat, frequency of known naturalisation in other geographical areas, geographic connectivity of species distribution, reproductive patterns and differentiated genetic diversity. These criteria represent a useful summary of potential lines of evidence. However, they do not represent an unambiguous decision-making framework and some of the criteria are operationally difficult to apply. For instance, assessment of native status on the basis of the type of habitat that a species currently occupies is difficult given the extent of human-induced habitat changes over time. In this article, we focus on the use of genetic data, in combination with historical and ecological evidence, to tackle this ‘native versus introduced’ question, focusing on field elms in the Balearic Islands.
The Balearic Islands belong to the same geological unit as the Baetic mountain range—in SE Spain—raised during the Alpine orogeny (Azanón et al., 2002). The islands maintained land connections with the Iberian peninsula along this range until the Pliocene marine transgression 5.2 Mya ago (Hsü et al., 1977). At the beginning of the Pliocene (5.32–5 Mya), the climate was more humid than today, and a strong change in climatic conditions took place from 3.2 to 2.3 Mya, with the establishment of xeric conditions that promoted the expansion of sclerophyllous forests in Mediterranean Basin (Suc, 1984; Fauquette et al., 1999). Strongly buffered by warm marine influences, changes in temperature and rainfall in the Quaternary glacial–interglacial oscillations had a lower impact on plant distributions in the Balearics at less extent than in continental environments, permitting the survival of thermophilous species from the Tertiary flora (see palynological review in Carrión et al., 2012). This climatic and geographical history of the Balearic Islands has led to the current presence of multiple plant taxa of diverse origins derived from the paleogeographic and climatic events from Tertiary and Quaternary periods (Médail and Diadema, 2009).
Overlaid on these natural determinants of the plant diversity of the region, the long-term interaction between humans and Mediterranean islands has resulted in frequent plant introductions and island systems are particularly susceptible to colonisation by non-native species (Hulme et al., 2008). Examples of invasive introduced species in the Balearic Islands include Araujia sericifera, Carpobrotus edulis and Oxalis pes-caprae (Sanz Elorza et al., 2004). In addition, several tree species that are widely distributed on the mainland have been deemed exotic when they are rare on the islands (for example, Quercus suber (Bonner, 1977); Populus alba and Fraxinus angustifolia (Bolòs and Molinier, 1969)). However, the use of rarity per se as a criterion for non-native status is confounded by natural rarity or that induced by the intense human transformation of Mediterranean landscapes. Of particular interest in this respect, is the extremely rare riparian forest in the Balearics, nowadays mostly represented by few, scattered, populations of field elm (Ulmus minor Mill.), confined to single lines of trees along the sides of the temporary watercourses of torrents in Majorca and only in ravines in Minorca. There is a progressive decline in pollen records for different temperate and riparian (including Ulmus) taxa since the mid-Holocene in the Balearic Islands (see data from Pérez-Obiol and Yll (2003) in Ninyerola et al., 2009). This decline in riparian vegetation likely due to aridification was further amplified by progressive intensification of human activity since 2000 BC (Pérez-Obiol and Sadori, 2007). Ulmus pollen disappeared from palynological records before 2000 YBP in Majorca and 900 YBP in Minorca (see pollen diagrams from Pérez-Obiol and Yll (2003) in Ninyerola et al., 2009; reviewed Balearic records in Carrión et al., 2012). Collectively, this information suggests that Ulmus was native to the islands, but the lack of continuous palynological records could be interpreted as evidence that the current populations are introduced.
The main historical large-scale human translocation of field elm was during Roman times, as the tree was used as structural support for grapevines (Fuentes-Utrilla et al., 2004). However, a previous chloroplast marker survey showed that the Romans did not introduced elms from Italy into the Balearic Islands (Gil et al., 2004). All historical references available from the early 1400s and onwards, depict Balearic elms alongside water course (Gil et al., 2003), that is, in their natural habitat. Consequently, if the elms are introduced it is likely to have been before 1400. In summary, field elms are rare in the Balearic Islands. The evidence suggests they were historically native, but it is possible that the current population is due to a pre-1400 (re)introduction. At present, the Balearic elms are not afforded conservation status and the species is considered introduced by some authors (Bolòs and Molinier, 1969; Moragues Botey and Rita Larrucea, 2005). To test this, we have used genetic markers (nine nuclear microsatellite loci) combined with ecological and historical information, following the principles established by Webb (1985). If the elms are native, we expect them to show high levels of genetic differentiation from the mainland populations and among the different islands. Any genetic affinities with mainland population should be strongest with the Baetic mountains in SE Iberia, which are part of the same geological unit as the Balearics. As field elms can spread clonally, if large clones are present, this would be consistent with native status, as clone size (at least to some degree) in tree species is indicative of long-term in situ persistence (Ennos, 2003). However, if the elms are introduced, we would expect to find low differentiation among populations and a subset of the variation found in the populations of origin, which are likely to be those in modern-day Catalonia and Rousillon, from where most of Christian settlers of the Balearics originated.
Materials and methods
Population sampling
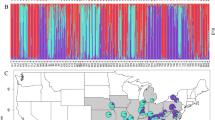
Nine populations of Ulmus minor were sampled during the spring and early summer of 2005 (Figure 1): three populations were collected in each of Majorca and Minorca islands and three in the Iberian peninsula aiming to represent regional genetic variability. The three populations from Majorca were selected in separated streams of the Tramuntana mountain range (Clot d'Aubarca (CAU), Son Brondo (SBR) and Sa Riera (SRI); Figure 1b), away from the east and the flat central part of the island deeply affected by Dutch Elm disease, what may have reduced the original genetic diversity. In Minorca, where Dutch Elm disease is absent, elm populations grow scattered in the few gullies and temporary streams that run mostly parallel to each other from the central part of the island to the North and South coasts. We chose two populations located in two gullies in the southern part of the island (Algendar (AGD) and Albranca (ABR)), where elms are more abundant, and one in a temporary stream in the northern part (Sant Joan des Horts (SJO)) (Figure 1c). The Iberian populations have been chosen as references for genetic diversity to track the putative origin of Balearic genetic variables in two different regions: (i) one population (Río Mundo (RMD)) in the Baetic mountains, which is the most diverse in terms of chloroplast DNA haplotypes (Gil et al., 2004) and where pollen records suggest the existence of a glacial refugium for the species (Carrión, 2002); (ii) two populations from Catalonia (Río Ter (RTR) and Río Gaià (RGA)) sampled in different river systems separated by >100 km and covering the distribution range of the species in this region. Pollen records also indicate a glacial refugium for Ulmus in NE Spain (González-Sampériz et al., 2005).
Geographic location of the nine populations of Ulmus minor analysed. (a) Overall location of the populations in the Iberian peninsula and the Balearic Isles. Detailed maps with the location of the populations from Majorca (b) and Minorca (c) are given. In these two maps, 1 × 1 km UTM squares with presence of Ulmus minor in each island are included (modified from data in BioAtles 2.0, Govern de les Illes Balears, http://bioatles.caib.es). Note that different elevation gradients have been used in the maps of the islands to highlight their geographic features.
Thirty trees per population were sampled, except in Majorca where only 21 individuals in CAU and 29 trees in SBR and SRI were collected. In total, 259 trees were sampled. Whenever possible, individuals were located at least 50 m far from each other to minimise sampling of clonal genotypes. Exact position of each population was determined using a global position system (GPS, Trimble, Geo XT, 2005).
DNA extraction and microsatellite amplification
DNA extraction was carried out following the protocol of Dumolin et al. (1995) with the following modifications: 1,4-diothiothreitol was used instead of β-mercaptoethanol and a small quantity of insoluble polyvinylpirrolidone was added before the addition of extraction buffer. After extraction, DNA was quantified and diluted to 5–10 ng μl–1 to use as template for PCR reactions.
Nine microsatellite loci were used for this study, six of them specifically developed for Ulmus minor (Ulmi1-11, Ulmi1-21, Ulmi1-98, Ulmi1-165, Ulmi2-16 and Ulmi2-20; Collada et al., 2004) and three microsatellite markers developed for U. laevis (Ulm2, Ulm3 and Ulm8; Whiteley et al., 2003). Polymerase chain reactions were carried out as described in Collada et al. (2004). Amplified PCR products were separated in 8% denaturing polyacrilamyde gels using a Li-Cor 4300 sequencer (Li-Cor Biosciences, Lincoln, NE, USA) and banding profiles scored using the software Gene ImagIR 3.56 (Scanalytics, Fairfax, VA, USA). Sequamark molecular size standard (Invitrogen, Carlsbad, CA, USA) was included in each gel to allow the calculation of allele sizes.
Clonal structure and genetic diversity analysis
Microsatellite data were analysed with GenAlEx v.6 macro in Excel (Peakall and Smouse, 2006). We first checked for the presence of repeated (clonal) multilocus genotypes (MLGs), verifying that the set of loci used permits as a good estimate of the real number of MLGs. To describe the clonal structure of each population, we reported several standard measures obtained with the program GENCLONE (Arnaud-Haond and Belkhir, 2007): (i) the number of samples (N), the number of observed MLGs (G), the number of repeated MLGs (GR), the number of unique MLGs (GU) and the number of locally private MLGs (GL); (ii) the modified index of genotypic richness R = (G–l)/(N–1) (R varies from 0, when all N plants in a population possess the same genotype, to 1.0, when all individuals present distinct genotypes); and (iii) the complement of Simpson’s index of diversity D=1–Σ(ni(1–ni)/N(1–N)), where ni is the number of trees of genotype i. We also used GENCLONE to test the clonal identity of repeated MLGs in each population: first, the genotype probability, Pgen, is calculated using population allele frequencies by a round-robin method. Then the probability of a repeated MLG being derived from distinct reproductive events, Psex, is calculated for each of the re-encounters (from n =2 to n = ni), which is used to test whether all identical replicates are actually clones. If Psex <0.05 from the first re-encounter (n =2), all the trees of that MLG were assigned to the same clone. When a repeated MLG showed a significant Psex for n >2, the number of ramets that were significant is reported. Owing to software restrictions to handle missing data, we eliminate locus Ulmi1-11 from the analyses because a large number of the samples presented null alleles. Similarly, two samples presenting null alleles in locus Ulmi2-16 were also excluded. However, these samples and locus Ulmi1-11 data were considered when the repeated MLGs were defined. Only one copy per MLG within a population was considered in all further analyses.
Standard genetic diversity indices (number of alleles, effective number of alleles and number of private alleles) were calculated for each population using GenAlEx. Furthermore, we used FSTAT v. 2.9.3.2 (Goudet, 2001) to estimate observed (HO) and expected (HE) heterozygosities, and the inbreeding coefficient (f) for each locus and overall loci in each population. Tests for heterozygote deficit and excess (departure from Hardy–Weinberg equilibrium) were performed by randomising alleles among individuals. P-value for the tests was based on 1640 randomisations and strict Bonferroni corrections were considered to obtain the adjusted nominal level for multiple comparisons, for an initial α of 0.05.
Genetic clustering of populations
To analyse the genetic structure of the sampled individuals, we first carried out a principal coordinates analysis (PCoA) based on pairwise, individual-by-individual genetic distance matrix using GenAlEx v.6 (Peakall and Smouse, 2006). The analysis was performed including and excluding locus Ulmi1-11.
We compared the results of the PCoA with two Bayesian approaches based on different inference methods for the detection of genetic clusters without geographic information (BAPS v.4.13 (Corander et al., 2006) and STRUCTURE v.2.0 (Pritchard et al., 2000)) to assess confidence in the results obtained (Latch et al., 2006). BAPS uses a stochastic optimisation method to infer the posterior mode of population structure, which results in much faster runs than the Markov chain Monte Carlo algorithm implemented in STRUCTURE. We performed a mixture analysis at group level in BAPS to analyse the clustering of populations, followed by an admixture analysis to identify the proportion of each individual’s genome assigned to the resulting genetic clusters. The most likely number of clusters (K) was obtained after 10 independent simulations for values of K ranging from 1 to 9. We ran STRUCTURE five times for each K ranging from 1 to 7 (any genetic structure was lost for K-values higher than 5) for 106 iterations and a burn-in period of 105. We used the admixture model with correlated allele frequencies as the populations hypothesised to have diverged from a common ancestral population; these priors improve clustering of closely related populations allowing to detect subtle genetic structure among groups (Falush et al., 2003). To facilitate selection of the most appropriate K value, we used the ΔK statistics (Evanno et al., 2005). We checked the effect of locus Ulmi1-11 by running the Bayesian analyses with it included and excluded from the data set.
Results
Genetic diversity within populations
The total number of alleles scored in the 259 individuals analysed with the nine microsatellites was 97, ranging from a minimum of 1 allele (Ulmi1-11) to a maximum of 25 (Ulmi2-20) with an average of 10.7 alleles per locus (see Appendix). Excluding locus Ulmi1-11 (that only presented PCR products in trees from Majorca), the Balearic populations had on average 50% less allelic richness (average A<3 with maximum A=4) than the Iberian populations (mean A=6.3, range 4–15 in RTR, and A=4–10 in RMD and RGA). Even RMD, the Iberian population with lower allelic richness (overall A=5.1), had twice as many alleles per locus than all the Balearic populations for most loci. All but two populations (SBR in Majorca and ABR in Minorca) presented private alleles, ranging from 1–3 in the Balearic populations to 3–12 in Iberia. However, if we consider the total number of alleles in each of these populations, the percentage of private alleles in the islands (6.25–12.5%) is comparable to that in the mainland (6.52–7.40%). Both positive and negative values of the inbreeding coefficient were observed in all loci, varying among populations. However, significant inbreeding was only detected in the Iberian populations for one or two loci; only RMD showed significant departure from Hardy–Weinberg equilibrium (P<0.05) across loci, mostly because of deviation in Ulmi2-16; if this locus is excluded the overall test for RMD is not significant.
Clonal structure and genotypic diversity
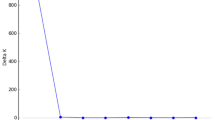
Among the 259 individuals analysed, the total number of detected MLGs with 8 of the 9 microsatellites used was 92 (G), distributed in 78 unique genotypes plus 14 repeated genotypes (GR). The addition of Ulmi1-11 did not modify the number of genotypes detected (Figure 2), which indicates that the number of microsatellites used was enough to accurately distinguish all different genotypes in the data set.
Multilocus matches and unique genotypes by locus for increasing combinations of the nine loci analysed. Solid line with black squares indicates number of samples with matching MLGs. Dashed line with white squares indicates number of samples with unique MLGs (GU). Values for each increasing combination of loci are indicated. The final 181 samples with matching genotypes were distributed across 14 different MLGs (GR).
We found contrasting levels of genotypic diversity and clone size (number of copies or ramets) per population in the Balearic Islands compared with the Iberian populations (Table 1). The genotypic richness R in the islands was low, with a maximum of 0.11 in SBR. The low values for R reflects the small number of MLGs observed in the populations from Majorca and Minorca, with a maximum of four genotypes in SBR and a minimum of one in two populations from Minorca (AGD and SJO). However, despite the low number of genotypes observed in Balearic populations, there was a considerable variation in Simpson’s diversity index, with D ranging from 0 to 0.65, as a result of differences in the number of ramets of repeated MLGs. For example, the diversity index of CAU (D =0.42) is higher than in SRI (D =0.14), despite the former having one less genotype (two versus three), because in CAU the two genotypes are more evenly shared among the trees, whereas in SRI, one genotype comprises 27 out of 29 trees. In general, the number of individuals belonging to each repeated MLG was high, ranging from four individuals up to the entire sampled population (Table 2). Interestingly, in both islands at least one repeated genotype appeared in two different populations. In Majorca, SBR shared genotype B with CAU, located 26 km apart from each other and genotype D with SRI, located 11 km apart from SBR. In Minorca, genotype F appeared in ABR and SJO, located 10 km apart from each other.
Tests for vegetative reproduction were significant (Psex <0.05) for most repeated genotypes. In only three MLGs (D in SRI, E in AGD, and F in SJO and ABR) the Psex-values were not significant from the first re-encounter, although they did show significance after the second or third re-encounter. As these MLGs constitute most of the trees in their respective populations (Table 2), their alleles appear at high frequencies in the population. This results in not only high genotype Pgen-values, but also in the somehow high probability of these repeated genotypes arising from independent sexual reproduction events (Psex-values), which explains why they are nonsignificant for the firsts re-encounters. Therefore, we considered all individuals from repeated MLGs D, E and F to have a clonal origin. Clone size within populations, estimated as the maximum distance between ramets of the same clone, varied from 152 m for clone G in ABR to over 1500 m for clone D in SRI. We found four single-copied MLGs (unique genotypes, GU) in the Balearic Islands, three in Majorca and one in Minorca (Table 2). Three of them (genotypes 2 and 3 in SRI and genotype 4 in ABR) presented genotypes only slightly different to the multilocus profiles of the most common clonal genotype in the population. We considered them to be the result of somatic mutations, as putative score errors were discarded by re-extracting and re-amplifying these samples and at least one sample of the original clone.
Contrary to the island populations, most of the Iberian peninsular trees showed different MLGs (Table 1). In each population, between one and four repeated genotypes (GR) were found, all of them of clonal origin (Psex <0.05 for the first encounter). All these clones consisted of two copies, except one clone in RMD with four ramets (clone size 176 m). Individuals belonging to the same clone had been sampled adjacent to each other (distance between ramets ranging 24–96 m). Genotypic richness in the mainland populations was much higher (R=0.86) than in the islands (R=0.11). Moreover, in all Iberian populations almost all MLGs were represented by a unique tree, resulting in Simpson’s diversity indexes close to 1.00.
Genetic clustering of populations
We used pairwise, individual-based genetic distances in a PCoA to investigate the relative position of genotypes in a multidimensional space. The first three principal axes using eight microsatellites accounted for 38.34%, 18.94% and 12.54% of the variation, (total variation explained=69.82%). The inclusion of locus Ulmi1-11, the marker depicting one single allele solely in Majorca, only slightly modified the variation explained by each axis (37.06%, 20.57% and 12.42%; 70.05% altogether).
The distribution of MLGs along axes showed an overall correlation with their geographic origin (Figure 3). Axis 1 differentiated the individuals of the Baetic mountains (RMD) from those of the Catalonian (RTR and RGA), leaving the Balearic MLGs between both groups. The samples from Majorca and Minorca were separated to a lesser extent by the first axis. Axis 2 separated the genotypes of Minorca and all but two genotypes from Majorca (MLGs B and C) from the Iberian ones. Finally, axis 3 separated the two Catalonian populations, although the separation is imperfect. When locus Ulmi1-11 was considered, a clear differentiation between the two islands appeared, except one genotype from Majorca that clustered within those from Minorca.
PCoA plots for the different MLGs of the nine Ulmus minor populations analysed. Plots show the distribution of the samples along the first, second and third axes. Percentage of variation explained by each axis is indicated in each plot. (a) PCoA plots considering the genetic data of all microsatellites but Ulmi1-11; (b) PCoA plots when all the nine microsatellites, including Ulmi1-11, are included in the analysis.
The Bayesian clustering analysis corroborated the results of the PCoA showing no differences with the inclusion or exclusion of locus Ulmi1-11. The analysis of population clustering with BAPS had K=5 as the most likely number of clusters (P=0.99883) among the nine populations, corresponding to each of the three Iberian populations and Majorca (CAU, SBR and SRI) and Minorca (AGD, SJO and ABR) (Figure 4a). Overall, the admixture procedure assigned each individual’s genome almost entirely to its original cluster, with only a noticeable degree of admixture observed between the Catalonian populations (RTR and RGA).
Results of Bayesian admixture analysis for all distinct MLGs of Ulmus minor in six Balearic populations at Majorca (CAU, SBR and SRI) and Minorca (AGD, SJO and ABR), one south-eastern Iberian population at Baetic mountains (RMD) and two north-eastern populations at Catalonia (RTR and RGA), using genetic data of nine nuclear microsatellites. In each graph, a column represents one MLG and the different colours represent the contribution of each genetic cluster to an individual genotype. High correspondence of genetic clusters with geographic populations indicates independent evolutionary stories in each region. (a) Results of admixture of genotypes for the five clusters detected with BAPS. (b) Results of admixture obtained with STRUCTURE for K =2, 3 and 4.
The individual-based clustering method implemented in STRUCTURE detected a smaller number of clusters. The value of ΔK identified K=2 as the most probable number of clusters, although K=3 also presented a high ΔK (Figure 5). The admixture analyses were coherent with the geographic location of the samples for K=2, 3 and also 4, with individuals of the nine populations clustered together according to different levels of genetic similarity (Figure 4b). Thus, for K=2, cluster one is formed by the individuals from the Balearic Islands together with those in Baetic mountains (RMD), and the Catalonian samples formed the other cluster. In the next level of clustering (K =3), the individuals of the previous cluster one were separated in two clusters, with the Balearic samples in one and the individuals from RMD in the other. Finally, considering K=4, the two Catalonian populations were separated, although in this case a certain degree of admixture between the populations was observed. The genomes of two individuals from Majorca and three from the Baetic mountains always showed certain similarity to a different cluster, although the degree of assignment was not consistent among analyses (K =2–4). No further recognisable population groups were observed with STRUCTURE for K >4.
Results of the Bayesian clustering of Ulmus minor individuals using STRUCTURE. (a) Mean posterior probability (L(K) ± s.d.) for each K value over five runs. (b) Evanno et al. (2005) statistic ΔK, showing a peak at the ‘true’ value of K (uppermost level of structure), here two clusters (but notice that K=3 also shows a high value for ΔK).
Discussion
Webb (1985) proposed eight criteria for determining species nativeness. In the absence of clear historical or habitat-related evidence, we present here the first study that uses genetic data to solve the controversy of whether field elm (Ulmus minor), a rare riparian tree in the Balearic Islands, is or is not native to the archipelago. To do this, we analysed the genetic structure of Balearic and Iberian populations in two different regions, one which belongs to the same geological unit (Baetic mountains) and the other (Catalonia) with strong historical connections with the islands.
Nativeness and ancient origin of Ulmus minor in the Balearic Islands
All three different clustering methods (PCoA, BAPS and STRUCTURE) showed high genetic differentiation of the Balearic genotypes compared with those from Iberia. Also, despite the smaller number of genotypes observed, the populations in the Balearic Islands had similar percentages of private alleles (per total number of alleles) to the Iberian populations. If Balearic elms were derived from the Catalonian populations, we would have expected to see more alleles shared between both regions. In contrast, our results point to a divergent evolution in each region with novel alleles arising in allopatry. For instance, 4 (36%) of the Balearic genotypes possess the Ulmi1-11 allele absent in all 81 Iberian genotypes.
A previous survey using genetic evidence to study human movement of elm (Gil et al., 2004) suggested massive propagation of Italian elms during Roman times. However, chloroplast markers indicate that the Italian trees (including Sicilian and Sardinian) were not present in the Balearics (Gil et al., 2004). In this study, the nuclear microsatellite data show that Balearic elm genotypes are clustered independently from Baetic and Catalonia populations, suggesting that natural patterns, rather than patterns caused by human introduction, are responsible for the observed variation.
Elms probably arrived in the Balearics during the Messinian salinity crisis, when the islands were linked to the Iberian peninsula through a land corridor connecting them with the Baetic mountains. The islands have remained isolated since the Pliocene marine transgression 5.2 Mya ago. STRUCTURE analyses clearly cluster the Balearic elms with the Baetic RMD trees for K=2, the most likely K-value (Figure 4b). When Catalonian populations are excluded from the analysis (data not shown), the Balearic elms always remain clustered together whereas Baetic genotypes consist of a differentiated group of admixed individuals from one to three genetic clusters (K=4 is the most likely K-value in this case). This grouping probably reflects the ancestral Baetic origin of the Balearic populations.
In the case of Majorca and Minorca, the two islands were connected during the Quaternary glaciations, including the last glacial maximum 18 000 bp, when sea level descended (Shackleton et al., 1984). Thus, each glaciation provided an opportunity for gene exchange between islands. Here, BAPS analysis clearly separates the genotypes of each island. This separation is also apparent, albeit with some overlap, along the third principal axis when Ulmi1-11 is considered in PCoA analysis. STRUCTURE keeps the insular genotypes together, pointing to a common ancestral origin. Therefore, current differentiation may have occurred during the last interglacial. However, if, as discussed below, the observed clonal structure is an ancient feature in the islands, the scarcity of sexual reproduction would have slowed down the differentiation process, in which case the genetic structure observed between islands probably predates the last glacial period.
Fossil evidence suggest the survival of Ulmus in many areas of NE Spain since the Pliocene (López, 2003; González-Sampériz et al., 2005). Although BAPS population-based clustering method is able to separate the two Catalonian populations (RTR and RGA), results from PCoA and STRUCTURE suggest that these populations have not undergone complete differentiation. Considering the geographic distance between both populations, current gene flow is unlikely to be responsible for the observed admixture. Alternatively, a recent splitting time could explain the lack of genetic divergence.
Clonal versus sexual reproduction in Ulmus minor
The Balearic populations of elms presented a small number of MLGs (11 overall, regardless of somatic mutations), most of them being large clones extending hundreds of metres. In a species able to reproduce both sexually and asexually, extended clones are to be expected where sexual regeneration is hindered (Steinger et al., 1996; Ennos, 2003). Vegetative reproduction is frequently observed in riparian plant species subject to natural cycles of disturbances like floods and droughts, which influence the proportion and distribution of sexual and asexual recruitment and therefore genotypic diversity levels among populations (Barsoum et al., 2004). The large differences in clonality observed between the Balearic and Iberian populations of Ulmus minor can thus be explained as the result of different environmental pressures related to the frequencies of disturbances affecting the sampled populations in past and present times.
The three Iberian populations grow along permanent watercourses, which experience regular floods after the snow melts in the spring, coinciding with the elm seed dispersal period. Spring floods open gaps in the vegetation creating bare land where new seedlings can establish. These conditions favour sexual recruitment, resulting in the high levels of genotypic diversity observed in the mainland rivers (Barsoum et al., 2004; López-Almansa, 2004). Conversely, the Balearic populations of elms grow scattered along channels of ephemeral streams fed by typical Mediterranean rainfall regimes, which maintain dry channels during several summer months and promote high and abrupt flows in autumn, with floods in the following months (Gasith and Resh, 1999). This pattern of rainfall reduces the opportunities for seed dispersal and seedling establishment. Furthermore, brief summer storms, sometimes very intense, often produce floods in the then dry streams. Established clones are much more likely to survive these violent discharges carrying tons of debris than newly established seedlings. At the same time, root fragments can be uprooted and transported in the flow, contributing to long distance recruitment of elm clones in the Balearic populations, similarly to what has been observed in other riparian species (Barsoum et al., 2004; Brundu et al., 2008; Macaya-Sanz et al., 2012). Given enough time, long distance dispersal of root fragments and vegetative propagation under benign climates could explain the presence of some clones in distant watersheds, albeit we cannot exclude human contribution to this feature (Brundu et al., 2008; Macaya-Sanz et al., 2012), or (despite our analyses suggesting otherwise), that these shared multi-locus genotypes between watersheds are independently sexually derived. Regardless, as somaclonal variations were not observed across the surveyed nuclear microsatellites in a 2000-year-old extensively propagated field elm clone (Gil et al., 2004), the presence of several somatic mutations in the Balearics points to very ancient clonal genotypes. The increasing xeric conditions of Balearic climate initiated 5000–6000 year BP (see reference in Ninyerola et al. 2009) could have allowed well-adapted clones to persist during millennia, contributing to the maintenance of established genotypes with well-developed root systems and therefore the marked clonal structure found. Furthermore, being one of the most palatable trees in the islands, elm are likely to have suffered strong browsing both before the arrival of the first humans (from the endemic and extinct goat Myotragus balearicus; Alcover and Bover, 2002) and later by domestic livestock. Seedlings are more easily destroyed by grazing that root suckers, whose vigorous growth allows them to exceed browse-line rapidly, thus favouring the vegetative spread of the clones along the streams.
Despite the small number of genotypes, seed set is frequently observed in populations from both Balearic islands (P Fuentes-Utrilla, unpublished data; P Fraga, personal communication). Ulmus minor is an anemophilous, outcrossing species in which autopollination is very rare (see López-Almansa, 2004). Although nowadays seedling recruitment seems to be prevented, and unless putative inbreeding depression would act at seedling stage, the fact that fully developed seeds are currently produced reveals the potential of the species to increase genotypic diversity under more suitable environmental conditions.
Implications for conservation
The Balearic elm populations are formed by a small number of genotypes that have been able to endure the drier conditions that have dominated the climate of the archipelago during the last 6000 years. The ability of Ulmus minor to stand summer drought was probably important in its survival compared with local extinction of less tolerant species like beech or alder (Ninyerola et al., 2009; Carrión et al., 2012). However, increasing water demand for tourism in the last decades has resulted in aquifer over-exploitation (Mediterranean_Groundwater_Report, 2007), and the resultant lowering of the water table may put additional stress on the existing trees, making them more vulnerable to the attack of pests and diseases.
The ‘aggressive’ strain of Dutch Elm disease arrived in the Balearic Islands via Majorca in the early 1990s, causing the death of hundreds of trees in Majorca and most trees in Ibiza (García Rotger and Romero Casado, 1996). Genetic results presented in this paper can now be considered in the Local Elm Conservation Action Plan (Florit García, 2005) to maximise the conservation of the Balearic Elm Genetic Resources: (i) the Balearic low genotypic diversity necessitates efforts to maintain most or all of the existing diversity; (ii) the clonal nature of the existing populations imply that the elimination of any population may significantly reduce the already low Balearic genotypic diversity; (iii) however, if the clonality turns out to be a general feature, a small number of ‘strategic ex situ populations’ could be established to contain all or most of the existing genetic variation. An extensive genotypic survey of the species across the Balearic Islands, maximising the number of localities sampled, would provide useful information to guide this sample selection process.
Conclusions
Whether a plant is native or alien to a certain region is too often based on opinion rather than on firm scientific evidence (Webb, 1985). No evidence was provided by Bolòs and Molinier (1969) to support their assumption that elms were introduced by man to the Balearic Islands. When arbitrary designations are uncritically assumed by other authors in technical documents, they can compromise the conservation of native species wrongly considered to be introduced. In this study, we provide evidence, which strongly supports Ulmus minor as being native to the Balearic Islands. Although the native status of other rare temperate forest trees (that is, Acer opalus subsp. granatense, Amelanchier ovalis, Sorbus aria, Ilex aquifolium and Taxus baccata) in similar relict conditions in the Balearics has never been questioned, some botanists have often regarded other temperate and riparian species (Corylus avellana, Fraxinus angustifolia and Populus alba) as introduced without a clear evidence base for their decision. In the light of our study here, following the precautionary principle, we suggest that, until genetic evidence of their introduction is provided, rare populations of hazelnut, ash and poplar should also be considered as native and included in conservation planning in the Balearic Islands.
Data archiving
Data available from the Dryad Digital Repository: doi:10.5061/dryad.bq4p7.
References
Alcover J, Bover P . (2002). Paleontología, espeleología y ciencias del karst en las Baleares. Bol. SEDECK 3: 92–105.
Arnaud-Haond S, Belkhir K . (2007). GENCLONE: a computer program to analyse genotypic data, test for clonality and describe spatial clonal organization. Mol Ecol Notes 7: 15–17.
Azanón J, Galindo-Zaldívar J, García-Dueñas V, Jabaloy A . (2002). Alpine tectonics II: Betic Cordillera and Balearic Islands. In: Gibbons W, Moreno M (eds) The Geology of Spain. Geological Society: London.
Barsoum N, Muller E, Skot L . (2004). Variations in levels of clonality among Populus nigra L. stands of different ages. Evol Ecol 18: 601–624.
Bolòs Od, Molinier R . (1969). Vue d'ensemble de la végétation des Îles Baléares. Plant Ecol 17: 251–270.
Bonner A . (1977) Plantes de les Balears.Editorial Moll. Palma de Mallorca.
Brundu G, Lupi R, Zapelli I, Fossati T, Patrignani G, Camarda I et al. (2008). The origin of clonal diversity and structure of Populus alba in Sardinia: evidence from nuclear and plastid microsatellite markers. Ann Botany 102: 997–1006.
Carrión JS . (2002). Patterns and processes of Late Quaternary environmental change in a montane region of southwestern Europe. Quat Sci Rev 21: 2047–2066.
Carrión JS, Fernández S, González-Sampériz P, López-Merino L, Peña L, Burjachs F et al. (2012) Paleoflora y Paleovegetación de la Península Ibérica e Islas Baleares: Plioceno-Cuaternario. Ministerio de Economía y Competitividad, Madrid http://www.paleodiversitas.org (last accessed January 2014).
Collada C, Fuentes-Utrilla P, Gil L, Cervera MT . (2004). Characterization of microsatellite loci in Ulmus minor Miller and cross-amplification in U. glabra Hudson and U. laevis Pall. Mol Ecol Notes 4: 731–732.
Corander J, Marttinen P, Mantyniemi S . (2006). A Bayesian method for identification of stock mixtures from molecular marker data. Fish B-NOAA 104: 550–558.
Dumolin S, Demesure B, Petit RJ . (1995). Inheritance of chloroplast and mitochondrial genomes in pedunculate oak investigated with an efficient PCR method. TAG 91: 1253–1256.
Ennos RA . (2003). The contribution of population genetic studies to plant conservation. Botanical J Scotland 55: 89–100.
Evanno G, Regnaut S, Goudet J . (2005). Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14: 2611–2620.
Falush D, Stephens M, Pritchard JK . (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587.
Fauquette S, Suc J-P, Guiot J, Diniz F, Feddi N, Zheng Z et al. (1999). Climate and biomes in the West Mediterranean area during the Pliocene. Palaeogeography Palaeoclimatology Palaeoecology 152: 15–36.
Florit García A . (2005) Resultats de l'inventari d'omedes de les Illes Balears. Conselleria de Medi Ambient. Govern de les Illes Balears, Palma de Mallorca, Spain. Informe inédito.
Fuentes-Utrilla P, López-Rodríguez RA, Gil L . (2004). The historical relationship of elms and vines. Invest Agrar: Sist Recur For 13: 37–45.
García Rotger M, Romero Casado J . (1996). Detección de la grafiosis agresiva en la isla de Ibiza (Baleares). Boletín de Sanidad Vegetal: Plagas 22: 789–801.
Gasith A, Resh VH . (1999). Streams in Mediterranean climate regions: abiotic influences and biotic responses to predictable seasonal events. Ann Rev Ecol Syst 30: 51–81.
Gil L, Fuentes-Utrilla P, Soto A, Cervera MT, Collada C . (2004). English elm is a 2,000-year-old Roman clone. Nature 431: 1053–1053.
Gil L, Manuel C, Díaz-Fernández P . (2003) La transformación histórica del paisaje forestal en las Islas Baleares, Madrid.
González-Sampériz P, Valero-Garcés BL, Carrión JS, Peña-Monné JL, García-Ruiz JM, Martí-Bono C . (2005). Glacial and Lateglacial vegetation in northeastern Spain: new data and a review. Quatern Int 140–141: 4–20.
Goudet J . (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Available from: http://www2.unil.ch/popgen/softwares/fstat.htm (last succesful accessed March 2007). Updated from: Goudet J, 1995. FSTAT (vers. 1.2): a computer program to calculate F-statistics. J Hered 86: 485–486..
Hsü KJ, Montadert L, Bernoulli D, Cita MB, Erickson A, Garrison RE et al. (1977) History of the Mediterranean salinity crisis 267: 399–403.
Hulme PE, Brundu G, Camarda I, Dalias P, Lambdon P, Lloret F et al. (2008). Assessing the risks to Mediterranean islands ecosystems from alien plant introductions. In: Tokarska-Guzik B, Brock JH, Brundu G, Child L, Daehler CC, Pyšek P. (eds) Plant Invasions: Human Perception, Ecological Impacts and Management. Backhuys Publishers: Leiden, The Netherlands. pp 39–56.
Latch EK, Dharmarajan G, Glaubitz JC, Rhodes OE . (2006). Relative performance of Bayesian clustering software for inferring population substructure and individual assignment at low levels of population differentiation. Conserv Genet 7: 295–302.
López-Almansa JC . (2004). Review. Reproductive ecology of riparian elms. Invest Agrar Sist Recur For 13: 17–27.
López RA . (2003). Paleobotánica de los olmos. In: Gil L, Solla A, Iglesias S. (ed) Los Olmos Ibéricos. Conservación y Mejora Frente a la Grafiosis. Organismo Autónomo Parques Nacionales: Madrid.
Macaya-Sanz D, Heuertz M, Alba N, López-de-Heredia U, de-Lucas AI, Hidalgo E et al. (2012). The Atlantic-Mediterranean watershed, river basins and glacial history shape the genetic structure of Iberian poplars. Mol Ecol 21: 3593–3609.
Médail F, Diadema K . (2009). Glacial refugia influence plant diversity patterns in the Mediterranean Basin. J Biogeogr 36: 1333–1345.
Mediterranean_Groundwater_Report. (2007) MEDITERRANEAN GROUNDWATER REPORT. Technical report on groundwater management in the Mediterranean and the Water Framework Directive. MED-EUWI WG ON GROUNDWATER.
Moragues Botey E, Rita Larrucea J . (2005) Els vegetals introduïts a les Illes Balears. Conselleria de Medi Ambient, Govern de les Illes Balears.
Ninyerola M, Sáez L, Pérez-Obiol R . (2009). Relating postglacial relict plants and Holocene vegetation dynamics in the Balearic Islands through field surveys, pollen analysis and GIS modelling. Plant Biosyst 141: 292–304.
Peakall R, Smouse PE . (2006). GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6: 288–295.
Pérez-Obiol R, Sadori L . (2007). Similarities and dissimilarities, synchronisms and diachronisms in the Holocene vegetation history of the Balearic Islands and Sicily. Veg Hist Archaeobot 16: 259–265.
Pérez-Obiol R, Sáez L, Yll E . (2003). Vestigis florístics postglacials a les Illes Balears i dinàmica de la vegetació holocènica. Organismes i Sistemes 18: 77–94.
Pritchard JK, Stephens M, Donnelly P . (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959.
Sanz Elorza M, Dana Sánchen ED, Sobrino Vesperinas E (eds). (2004) Atlas de las Plantas Alóctonas Invasoras en España, Madrid.
Shackleton JC, van Andel TH, Runnels CN . (1984). Coastal paleogeography of the central and western Mediterranean during the last 125,000 years and its archaeological implications. J Field Archaeol 11: 307–314.
Steinger T, Körner C, Schmid B . (1996). Long-term persistence in a changing climate: DNA analysis suggests very old ages of clones of alpine Carex curvula. Oecologia 105: 94–99.
Suc J-P . (1984). Origin and evolution of the Mediterranean vegetation and climate in Europe. Nature 307: 429–432.
Webb DA . (1985). What are the criteria for presuming native status? Watsonia 15: 231–236.
Weir BS, Cockerham CC . (1984). Estimating F-Statistics for the analysis of population structure. Evolution 38: 1358–1370.
Whiteley RE, Black-Samuelsson S, Clapham D . (2003). Development of microsatellite markers for the European white elm (Ulmus laevis Pall.) and cross-species amplification within the genus Ulmus. Mol Ecol Notes 3: 598–600.
Acknowledgements
We thank Luis Berbiela and Pere Fraga for their support in sampling Balearic elm populations, M Burón Barrio and RA López for helping collecting the material, and C Collada and MT Cervera for guidance in the lab. EA Gill made valuable comments on the manuscript. We thank Pete Hollingsworth for his valuable editing aid. This work was supported by the Dirección General para la Biodiversidad (MMARM, Spain). PF-U holded a FPI PhD grant co-funded by the Consejería de Educación (Comunidad de Madrid) and the European Social Fund. In memoriam of M Burón Barrio, to whom the Spanish Elm Conservation and Breeding Programme owes so much.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Appendix
Appendix
Genetic diversity indices for the nine populations of Ulmus minor studied. Number of alleles (A), effective number of alleles (Ae), number of private alleles (Ap), expected (HE) and observed (HO) heterozygosities, and inbreeding coefficient (f) according to Weir and Cockerham (1984). Values in bold for f indicate samples which deviate significantly from Hardy–Weinberg equilibrium (P<0.05) after sequential Bonferroni corrections (tests for heterozygote deficit or excess). Calculations are based on genotype-level analyses (G=the number of distinguishable MLGs in each population; note that several populations consist of only one single genotype; see Table 1).
Rights and permissions
About this article
Cite this article
Fuentes-Utrilla, P., Valbuena-Carabaña, M., Ennos, R. et al. Population clustering and clonal structure evidence the relict state of Ulmus minor Mill. in the Balearic Islands. Heredity 113, 21–31 (2014). https://doi.org/10.1038/hdy.2014.21
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2014.21
This article is cited by
-
Reproductive strategy of a temperate canopy tree Tilia cordata Mill. (Malvaceae) is related to temperature during flowering and density of recent recruits
Tree Genetics & Genomes (2022)
-
Genotyping ex situ trees of Abies nebrodensis translocated from the original Sicilian population to enrich the gene pool
Journal of Forestry Research (2022)