Abstract
Comparative genomic studies have revealed that mammals typically possess two or more tandemly duplicated copies of the α-globin (HBA) gene. The domestic rabbit represents an exception to this general rule, as this species was found to possess a single HBA gene. Previous electrophoretic surveys of HBA polymorphism in natural populations of the European rabbit (Oryctolagus cuniculus) revealed extensive geographic variation in the frequencies of three main electromorphs. The variation in frequency of two electromorphs is mainly partitioned between two distinct subspecies of European rabbit, and a third is restricted to the hybrid zone between the two rabbit subspecies in Iberia. Here we report the results of a survey of nucleotide polymorphism, which revealed HBA copy number polymorphism in Iberian populations of the European rabbit. By characterizing patterns of HBA polymorphism in populations from the native range of the European rabbit, we were able to identify the specific amino-acid substitutions that distinguish the previously characterized electromorphs. Within the hybrid zone, we observed the existence of a second HBA gene duplicate, named HBA2, that mostly represents a novel sequence haplotype, which occurs in higher frequency within the hybrid zone, and thus appears to have arisen in hybrids of the two distinct subspecies. Although this novel gene is also present in other wild Iberian populations, it is almost absent from French populations, which suggest a recent ancestry, associated with the establishment of the post-Pleistocene contact zone between the two European rabbit subspecies.
Similar content being viewed by others
Introduction
Comparative genomic studies have revealed extensive variation in the size and membership composition of the α- and β-globin gene families in mammals (Hoffmann et al., 2008a, 2008b; Storz et al., 2008; Opazo et al., 2008a, 2008b, 2009). In principle, studies of gene copy number variation within species could provide insights into the evolutionary forces that have shaped the observed copy number variation among species. However, with the exception of the thalassaemia blood pathologies in humans (which are associated with deletions of adult α- or β-globin genes; Higgs, 2001), there is very little information about globin gene copy number polymorphism in other species.
Comparative studies of the mammalian globin gene clusters have revealed an unusual history of lineage-specific gene duplication and deletion in the α-globin gene cluster of the European rabbit (Oryctolagus cuniculus) (Cheng et al., 1986, 1987; Cheng and Hardison, 1988; Hoffmann et al., 2008a). Specifically, the α-globin gene cluster of rabbit underwent a dramatic expansion by means of an en bloc triplication of a four-gene set that originally comprised two tandemly duplicated ζ-globin genes and single copies of the α- and θ-globin genes (5′-ζ-ζ-α-θ-3′). A functional α-globin gene (HBA) appears to have been retained in only one of the triplicate gene sets. Thus, although the majority of mammalian species possess at least two tandemly duplicated HBA paralogs, the rabbit appears to possess a single HBA gene (Cheng et al., 1986, 1987). As this information was based on genomic sequence data from the domesticated rabbit, and as domesticated rabbits represent a small fraction of all genetic diversity present in the species as a whole (Ferrand and Branco, 2007; Carneiro et al., 2011), surveys of natural populations are required to determine whether the unduplicated, single-gene state is fixed in rabbits, or whether there is population-level variation in HBA copy number.
The European rabbit comprises two distinct subspecies that are parapatrically distributed across the Iberian Peninsula: Oryctolagus cuniculus algirus, present in the southwest of Iberia, and Oryctolagus cuniculus cuniculus, which occurs in the northeast of Iberia (Biju-Duval et al., 1991; Branco et al., 2000). The two subspecies diverged in allopatry roughly 2 million years ago during the Quaternary glaciations (Biju-Duval et al., 1991; Geraldes et al., 2006) and have since come back into secondary contact, establishing a hybrid zone in central Iberia (Branco et al., 2002; Geraldes et al., 2005; Ferrand and Branco, 2007). Both hybrid and parental Iberian populations have been extensively characterized by different types of markers, such as allozymes (Ferrand and Branco, 2007), mitochondrial DNA (mtDNA) (Branco et al., 2000, 2002) and X- and Y-chromosomes (Geraldes et al., 2006, 2008), that are concordant in placing the centre of the hybrid zone in the central region of Iberia, though not associated to any ecological or physical transition. A detailed analysis of genetic variation across the hybrid zone showed that the width of the hybrid zone greatly varies between genetic markers, and that pure parental phenotypes are only found outside the hybrid zone, suggesting that the European rabbit hybrid zone is a tension zone (Barton and Hewitt, 1985; Carneiro et al., unpublished).
Electrophoretic surveys of HBA polymorphism in wild populations of European rabbits revealed the presence of five electromorphs, three of which are common and geographically widespread (HBA*1, HBA*2 and HBA*3) and two of which are relatively rare and geographically restricted (HBA*4 and HBA*5) (Ferrand and Branco, 2007; Campos et al., 2007). A sixth electromorph, HBA*6, was only observed in domestic rabbit (Campos et al., 2007). Two of the three most common electromorphs are strongly associated with alternative mtDNA lineages that are diagnostic of the two subspecies: HBA*1 is associated with mtDNA lineage B, found mostly in animals from the subspecies cuniculus, and HBA*2 is associated with mtDNA lineage A, characteristic of the subspecies algirus (Campos et al., 2007). In contrast, the third most common electromorph, HBA*3, is mainly found in individuals from the hybrid zone and shows no association with any of the mtDNA lineages (Campos et al., 2007, 2008). This latter result suggests that HBA*3 could represent a novel sequence variant that originated within the European rabbit hybrid zone. Novel allelic variants associated with hybrid zones, named ‘hybrizymes’, are a general phenomenon that have been reported for other nuclear loci in hybrid zones of several vertebrate species (Woodruff, 1989; Bradley et al., 1993; Hoffman and Brown, 1995; Schilthuizen et al., 1999, 2004; Godinho et al., 2006). Using a multilocus analysis, the abrupt geographic shifts in allele frequencies across the Iberian Peninsula suggest that the HBA polymorphism may have experienced a history of spatially varying selection (Campos et al., 2008) and adds further support to the hypothesis that HBA*3 could represent an ‘hybrizyme’.
Here we report a survey of nucleotide variation and copy number polymorphism in the adult HBA genes in wild European rabbit populations from the two subspecies and the hybrid zone. By cloning and sequencing the HBA genes, we were able to identify the specific amino-acid mutations that distinguish the previously characterized electromorphs. We also discovered the presence of a de novo HBA gene duplication that likely originated in the hybrid zone between O. c. algirus and O. c. cuniculus in the Iberian Peninsula.
Materials and methods
Sampling
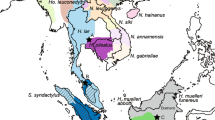
Using blood and tissue samples of rabbits that were sampled from each of the localities shown in Figure 1, we extracted DNA according to the standard salt protocols or by using the QIAGEN extraction kits, QIAamp Blood Kit and DNeasy Tissue Kit (Qiagen, Hilden, Germany). Population samples were grouped to represent the two native areas of the European rabbit subspecies, South-west Iberian peninsula (SWIP) and North-east Iberian peninsula (NEIP), the hybrid zone between the two subspecies (central Iberian peninsula, CIP) and the recently colonized areas, represented by French populations (see Figure 1 for details). Rabbits from the Portuguese islands of Flores (Azores) and Porto Santo (Madeira) are from the subspecies O. c. algirus (Fonseca, 2006) and were thus placed in the South-west Iberian peninsula group.
Sampling localities of the European rabbit populations and geographic groups defined for the analysis. The South-west Iberian peninsula (SWIP) populations (subspecies O. c. algirus) are: Cabreira (Cab), Portimão (Prt), Nisa (Ni), Seville (Sev), Doñana (Don), Huelva (Hue), Vila Viçosa (Vv), Santarém (San), Infantado (Inf) and Idanha (Id). The central Iberian peninsula (CIP) populations (from the contact zone) are: Bragança (Bra), Toledo (Tol), Albacete (Alb), Benavente (Bnv), Las Amoladeras (Ll), Ciudad Real (Cre) and Alicante (Alt). The North-east Iberian peninsula (NEIP) populations of O. c. cuniculus are: Lérida (Lle), Tarragona (Tar), La Rioja (Lrj), Zaragoza (Za) and Navarra (Nav). The French (FR) populations of O. c. cuniculus are: Perpignan (Per), Camargue (Tv), Vaulx-en-Velin (Vau), Carlucet (Car) and Versailles (Ver). Flores is an island in the Azores archipelago and Porto Santo is an island in the Madeira archipelago. The sampling localities that were only included in the survey of HBA copy number polymorphism (Figure 3) are denoted by a black dot. Pie charts indicate the relative frequencies of the HBA electromorphs. The electromorph HBA*6 was only observed in one domestic breed and in the population sample from the Tunisian island Kuriat and therefore is not represented in this figure.
PCR, cloning and sequencing of the HBA gene
All primers were designed by referring to the publicly available domestic rabbit HBA sequence (GenBank accession number M74142) (Hardison et al., 1991) and are provided in additional file 1. To identify the amino-acid mutations that distinguish the alternative HBA electromorphs, the complete coding region, and upstream and downstream flanking regions were analysed in specimens with known electrophoretic phenotypes. Genomic DNA (60–100 ng) was PCR amplified in a 25-μl mixture containing 2.25 μl of Buffer 10 × (Goldstar; Eurogentec, Liège, Belgium), 3 mM MgCl2, 0.8 mM dNTP's, 0.2 μl of primers HBA9B (M74142 nt 6303–6332) and HBA10C (M74142 nt 7544–7570), 10% DMSO and 2 U Taq DNA polymerase (Goldstar; Eurogentec). In an initial screening of nucleotide diversity, PCR products were purified using exonuclease and shrimp alkaline phosphatase (ExoSAP-IT; Amersham Biosciences, Fairfield, CT, USA), and were sequenced with the Big Dye Terminator Cycle Sequencing protocol on the ABI Prism 310 automated sequencer (Perkin-Elmer, Applied Biosystems, Carlsbad, CA, USA). Additional internal primers were also used in the sequencing of HBA (additional file 1). Because the sequences obtained through direct PCR were characterized by unusually high levels of polymorphism, particularly in the 3′-flanking region (see Results), we cloned a set of samples chosen to represent each of the four population groups and the five electrophoretic variants that segregate in those populations. PCR products were first cleaned with kit MinElute PCR Purification (Qiagen) and ligated mixtures were transformed into competent cell with the cloning kit pGEM-T Easy Vector System II (Promega, Madison, WI, USA), following the manufacturer's instruction. Colonies were grown overnight at 37 °C in solid LB medium with ampiciline, X-gal and IPTG. Recombinant plasmids were isolated with QIAprep Spin Miniprep kit (Qiagen) and inserts were confirmed with M13 primers using the PCR protocol developed to the direct amplification of HBA gene. Clones were sequenced using the Stabvida sequencing facilities (http://www.stabvida.com, Caparica, Portugal), with either M13 or specific HBA primers. Five clones per sample were initially sequenced. Because of the high number of polymorphic positions observed in the Iberian samples, additional sequences were obtained in order to unequivocally resolve all haplotypes. All mutations identified in the clones were confirmed by independent sequencing of at least two clones or by means of comparison with sequences obtained through direct sequencing of the PCR products. For comparative purposes, we also cloned and sequenced the HBA gene from the Eastern cottontail (Sylvilagus floridanus) and we annotated the HBA gene in the genome assembly of the American pika (Ochotona princeps) (GenBank accession number AC234618). We also used the published nucleotide (mRNA only) and amino-acid sequences of HBA gene from the plateau pika (Ochotona curzoniae) (GenBank accession number EF429202).
Population-level analysis of the three main allelic classes using PCR-RFLP
We developed a two-step PCR-RFLP protocol to distinguish the three common HBA electromorphs, HBA*1, HBA*2, and HBA*3. First, we followed the protocol of Ferrand et al. (2000), which discriminates electromorph HBA*1 from HBA*2 and HBA*3 using the restriction enzyme Sau96I. We then used a second enzyme, SmlI (New England Biolabs, Ipswich, MA, USA), which distinguishes electromorph HBA*2 from both HBA*1 and HBA*3. Enzymatic digestions with SmlI were conducted in accordance with the manufacturer's instructions and the DNA was separated in a 3% high-resolution agarose gel (NuSieve; Cambrex, East Rutherford, NJ, USA) and visualised using a solution of 0.04% ethidium bromide under UV light. Marker 5 (Eurogentec) was used to determine the approximate fragment sizes.
Population-level analysis of the HBA gene duplication
The population distribution and abundance of the HBA gene duplication was evaluated using a PCR-RFLP protocol based on two nucleotide substitutions in the 3′-flanking sequence that distinguish the HBA1 and HBA2 paralogs. A 313/312-bp fragment was amplified using the primers 10C and 3B (M74142; nt 7258–7275). The enzyme Hpy188I (New England Biolabs) was used in accordance with the manufacturer's instructions but with the inclusion of 10% DMSO in the reaction mixture. DNA was digested for 4 h at 37°C, separated by electrophoresis in a T9C5 polyacrylamide gel and visualized with silver staining. In each population sample, the frequency of the chromosome harbouring a single copy of the HBA1 or the HBA2 gene and the tandemly duplicated HBA1+HBA2 gene pair was calculated from the direct count of the observed PCR/RFLP genotypes.
The expectation maximization algorithm (Excoffier and Slatkin, 1995) was used to estimate the frequency of HBA gene-electromorph haplotypes. Standard error measures were obtained after 5000 bootstrap replicates. This analysis was done using Arlequin v3.0 (Excoffier et al., 2005).
Sequence analysis
The scaled recombination rate 4Nr (Hudson, 1987) and the number of intragenic recombination events in the history of the sample, Rm (Hudson and Kaplan, 1985), were estimated using the DnaSP 4.0. program (Librado and Rozas, 2009). The ϕw test, implemented in PhiPack (Bruen et al., 2006), is robust to the influence of homoplasy. This analysis was conducted using the default parameters and the significance of ϕw was obtained with 10 000 permutations. The recombination analysis was done excluding the two polymorphic microsatellites existing in this genomic region. A median-joining network (Bandelt et al., 1999) of all European rabbit HBA1 and HBA2 gene haplotypes was reconstructed using NETWORK 4.5.1.0 (Fluxus Technology LTD, Suffolk, UK). Statistical support for each node in the tree was obtained by bootstrapping (5000 replicates). All measures of nucleotide variability were calculated separately for each gene, HBA1 and HBA2. The divergence between the two European rabbit HBA genes and the American cottontail HBA gene was estimated using the raw and net divergence (Dxy and Da; Nei, 1987).
Results
Amino-acid differences that distinguish the alternative HBA electromorphs
By cloning and sequencing the HBA gene(s) in rabbits with known electrophoretic phenotypes (EMBL accession numbers HE608516–HE608566), we found that the HBA*1 electromorph is defined by the three-site amino-acid combination 29Val–48Phe–49Thr, the HBA*2 electromorph is defined by 29Leu–48Leu–49Ser and the HBA*3 electromorph is defined by 29Leu–48Leu–49Thr (see additional file 2 for a full compilation of all observed amino-acid mutations). This information allowed us to develop a PCR-RFLP protocol to distinguish the alternative alleles at the DNA level.
Patterns of nucleotide polymorphism and HBA copy number polymorphism
The direct sequencing of genomic DNA in some samples revealed a high heterogeneity in the 3′-flanking region in a pattern that suggested the existence of multiple HBA gene copies (additional file 3A). This pattern was absent in the two domestic rabbits we sequenced (additional file 2), confirming the previously documented absence of HBA gene duplication in domestic rabbits (Cheng et al., 1986). To investigate possible copy number variation in further detail, we sequenced an average of five independently cloned PCR amplicons per individual (range=3–9 clones per individual). To control for artifactual sequence changes produced by the cloning procedure, we only considered variants that appear in at least two independently cloned amplicons. In total, we obtained 68 sequences that allowed the identification of 51 unique haplotypes based on 54 nucleotide substitutions, three indels and two polymorphic microsatellites (additional file 4). In several samples, more than two sequences were recovered, a result that is compatible with the existence of at least one additional HBA gene (additional files 3 and 4). Two groups of sequences were observed and were defined based on a G7522T substitution and a single base deletion 7533delC (additional files 3B and 4). These mutations were attributed to two distinct HBA genes, named HBA1 (the previously described gene) (Cheng et al., 1986) and HBA2 (the newly discovered duplicate copy), respectively. The HBA2 paralog is further associated with the presence of more than 10 repeats of the mononucleotide C, located 31 nucleotides downstream from the stop codon (additional files 3B and 4). From this point onward the electrophoretic alleles will be distinguished from the gene paralogs by using an asterisk; for example, HBA*1 allele, HBA1 gene.
Of the 51 haplotypes observed, 34 were referable to the HBA1 paralog and 17 were referable to HBA2 (additional file 4). Relative to the HBA2 paralog, HBA1 is characterized by a higher level of nucleotide diversity (Table 1) and has experienced a greater number of intragenic recombination events (Table 2). The ϕw statistic (Bruen et al., 2006) for recombination in both HBA genes indicates the occurrence of a significant level of intralocus recombination (P<0.001), but does not exclude the combined occurrence of recurrent mutation.
Remarkably, HBA1 and HBA2 were segregating the same three major alleles (HBA*1, HBA*2 and HBA*3) because of a history of interparalog gene conversion (Table 3 and Figures 2 and 4). The history of gene conversion is also reflected by the fact that nucleotide divergence between the HBA1 and HBA2 paralogs of the European rabbit was much lower than the divergence between either of them and the single copy HBA gene in a closely related species, the Eastern cottontail (S. floridanus; additional file 5).
Distribution of the duplicate HBA genes and the spatial overlap with the three main electromorphs
A total of 303 animals from 24 Iberian (including two insular populations) and five French populations were screened for the presence of the HBA gene duplication by using the PCR-RFLP protocol (Figure 3). The survey revealed that this duplication is almost absent in French rabbit populations but is common in Iberian populations. Three additional domestic rabbits (CD PENA 15 and 55, breed Fauve Bourgogne and CD PENA 83, breed English) were also screened and in each case a single HBA1 gene was recovered.
Geographic distribution of chromosomes that harbour HBA1 or HBA2 gene in single copy and of chromosomes that harbour both paralogs simultaneously (HBA1+HBA2). The short names of populations are according with Figure 1. Numbers refer to the number of chromosomes surveyed per locality.
The gene duplication is not exclusively restricted to any particular subspecies or geographic region. However, chromosomes harbouring the duplicated HBA1–HBA2 gene pair were present at higher frequency in the zone of secondary contact (central Iberian peninsula, CIP; Figure 1). Also, the HBA2 gene was also observed in high frequency in single copy within the hybrid populations. Although the three main alleles were shared between the HBA1 and HBA2 paralogs, the HBA*3 allele is mainly associated with the HBA2 gene (Table 1; additional file 5).
Discussion
Differentiation between the HBA alleles
The overall levels of nucleotide polymorphism at the HBA locus are very high (Table 1), consistent with high levels of polymorphism observed at other nuclear genes in the European rabbit (Geraldes et al., 2006; Ferrand and Branco, 2007). By cloning and sequencing the HBA genes in specimens with different electrophoretic phenotypes, we were able to identify the specific amino-acid mutations that distinguish the three major electromorphs HBA*1, HBA*2 and HBA*3 (additional file 2). Based on contemporary patterns of allele frequency variation, the HBA*1 allele appears to trace its origins to the northeast of Iberia, within subspecies cuniculus, whereas the HBA*2 allele appears to trace its origins to the southwest, within subspecies algirus (Figure 1). The expansion of each group led to the establishment of the secondary contact zone between the two subspecies. Hybridization between the two rabbit subspecies may have provided the opportunity for recombination between distinct genetic backgrounds, giving rise to the recombinant allele HBA*3. Alternatively, allele HBA*3 could have originated through a point mutation in the parental allele HBA*2 (Figure 4) due to the putative increased mutation rates in hybrids (Hoffman and Brown, 1995; Schilthuizen and Gittenberger, 1994). Nevertheless, regardless of the precise mechanism underlying the origin of HBA*3, its prevalence in populations from the centre and surroundings of the hybrid zone (for example, Ciudad Real, Toledo and Idanha; Figures 1 and 3; Geraldes et al., 2008) is a clear indication that the origin of allele HBA*3 is associated with the post-pleistocene secondary contact of both rabbit subspecies.
Model for the origin of the HBA*3 allele, the HBA duplication and the copy number polymorphism observed in the European rabbit. (a) Allele HBA*3 originated in rabbits of hybrid origin, either by recombination between the parental alleles HBA*1 and HBA*2 (left) or by a mutation (*) in parental allele HBA*2 (right) and rose in frequency within the hybrid zone. (b) Unequal crossing-over between two chromosomes carrying the HBA*3 allele originated the HBA1–HBA2 gene duplication. (c) Subsequent unequal crossovers explain the existence of chromosomes carrying different number of HBA gene copies.
A model for the evolution of the HBA genes in the European rabbit
Recent studies have highlighted the fast rate of gene gain and loss in the multigene families of many mammalian species (Demuth et al., 2006; Hahn et al., 2007; Hoffmann et al., 2010). The pattern of gene turnover has been especially well characterized in the α-globin gene cluster of mammals (Zimmer et al., 1980; Hoffmann et al., 2008a; Storz et al., 2008).
Electrophoretic surveys of genetic variation at the HBA locus in natural and domestic populations of the European rabbit had previously revealed a pattern that was consistent with the existence of a single HBA gene, as reported by Cheng et al. (1986). However, our surveys of sequence variation at the HBA gene of wild European rabbits revealed the existence of multiple sequences in the 3′-flanking region, which suggested the presence of at least one additional HBA paralog. The HBA2 gene is nearly absent in France (we observed the duplication in only 1 out of 63 individuals; Figure 3), and in Iberia the HBA1–HBA2 gene duplication predominates in populations from the hybrid zone, where the allele HBA*3 also occurs at higher frequencies (for example, Ciudad Real, Toledo and Albacete; Figures 1 and 3). Since the domestication of the rabbit was a single event and all domestic rabbits originated from French populations (Ferrand and Branco, 2007; Carneiro et al., 2011), the reported absence of the HBA duplication in domestic rabbits (Cheng et al., 1986) is most likely a common feature of all breeds. This is consistent with surveys of HBA variation in New Zealand White rabbits (n=24) (Cheng and Hardison, 1988) and in three European breeds (English, Belgian Hare and Fauve de Bourgogne). Finally, the novel HBA2 gene mostly segregates the HBA*3 allele (Table 3), which explains why the HBA gene duplication was not detected in previous electrophoretic surveys. Taken together, these data suggest that the origin of the HBA duplication in the European rabbit is associated with the hybridization between O. c. algirus and O. c. cuniculus and with the emergence of the allele HBA*3, and that this event occurred within the European rabbit hybrid zone. Thus, the duplication originated sometime in the last 2 million years. The introgression of chromosomes harbouring the HBA1–HBA2 duplication provides further opportunities for unequal crossing-over to produce new gene arrangements (Figure 4).
Given that the HBA gene is present as a single copy in the Eastern cottontail and in pikas, it appears that this gene was present in an unduplicated, single-copy state in the common ancestor of lagomorphs. This would mean that the HBA1–HBA2 duplication is a derived condition specific to O. cuniculus in central Iberia, and thus restores the typical two-gene arrangement observed in most other mammalian species. As the HBA genes encode subunit polypeptides of adult haemoglobin, duplication and divergence of globin genes could potentially expand the repertoire of functionally distinct haemoglobin isoforms, as documented for the β-chain haemoglobins of mice (Storz et al., 2007, 2009, 2010; Runck et al., 2009, 2010). Spatially varying selection has been invoked to explain the geographic distribution of HBA alleles (Campos et al., 2008), which may also reflect the dissimilar distribution in the number of HBA gene copies. It is not clear whether variation in the copy number of HBA genes in the European rabbit has any fitness consequences. Although changes in gene dosage are generally expected to have deleterious effects, copy number polymorphism may also represent an important source of adaptive regulatory variation.
In conclusion, our work describes the remarkable geographic concordance between a new allelic variant, HBA*3, and a new HBA gene copy, HBA2, within the hybrid zone of the European rabbit. Although a more detailed analysis of the HBA gene cluster in hybrid individuals is still necessary to properly characterize this novel genetic combination, our results highlight the relevance of hybrid zones as highly dynamic sources of genetic novelties. Finally, the recent HBA duplication within natural populations of the European rabbit adds to the growing appreciation of the prevalence of copy number polymorphism in natural populations.
Data archiving
Sequences deposited in the EMBL repository: O. cuniculus HBA1 and HBA2 gene accession numbers HE608516 and HE608566; Sylvilagus cuniculus HBA gene accession number HE608842.
Data are deposited in the Dryad repository: doi:10.5061/dryad.j2p4d5t4
Accession codes
Accessions
GenBank/EMBL/DDBJ
References
Bandelt H-J, Forster P, Röhl A (1999). Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16: 37–48.
Barton NH, Hewitt GM (1985). Analysis of hybrid zones. Annu Rev Ecol Syst 16: 113–148.
Biju-Duval C, Ennafaa H, Dennebouy N, Monnerot M, Mignotte F, Soriguer RC et al. (1991). Mitochondrial DNA evolution in lagomorphs: origin of systematic heteroplasmy and organization of diversity in european rabbits. J Mol Evol 33: 92–102.
Bradley RD, Bull JJ, Johson AD, Hillis DM (1993). Origin of a novel allele in a mammalian hybrid zone. Proc Natl Acad Sci USA 90: 8939–8941.
Branco M, Ferrand N, Monnerot M (2000). Phylogeography of the European rabbit (Oryctolagus cuniculus) in the Iberian Peninsula inferred from RFLP analysis of the cytochrome b gene. Heredity 85: 307–317.
Branco M, Ferrand N, Monnerot M, Templeton AR (2002). Postglacial dispersal of the European rabbit (Oryctolagus cuniculus) on the Iberian Peninsula reconstructed from nested clade and mismatch analyses of mitochondrial DNA genetic variation. Evolution 56: 792–803.
Bruen TC, Philippe H, Bryant D (2006). A simple and robust statistical test for detecting the presence of recombination. Genetics 172: 2665–2681.
Campos R, Branco M, Weiss S, Ferrand N (2007). Patterns of hemoglobin polymorphism [α-globin (HBA) and β-globin (HBB)] across the contact zone of two distinct phylogeographic lineages of the European rabbit (Oryctolagus cuniculus).. In: Weiss S, Ferrand N. (eds). Phylogeography of European Refugia. Springer: Netherlands. pp 237–255.
Campos R, Storz JF, Ferrand N (2008). Evidence for contrasting modes of selection at interacting globin genes in the European rabbit. Heredity 100: 602–609.
Carneiro M, Afonso S, Geraldes A, Garreau H, Bolet G, Boucher S et al. (2011). The genetic structure of domestic rabbits. Mol Biol Evol 28: 1801–1816.
Cheng JF, Hardison RC (1988). The rabbit α-like globin gene cluster is polymorphic both in the sizes of BamHI fragments and in the numbers of duplicated sets of genes. Mol Biol Evol 5: 486–498.
Cheng JF, Raid L, Hardison R (1986). Isolation and nucleotide sequence of the rabbit globin gene cluster ψξ-α1-ψα. Absence of a pair of α-globin genes evolving in concert. J Biol Chem 261: 839–848.
Cheng JF, Raid L, Hardison R (1987). Block duplication of a ζ-ζ-α-θ gene set in the rabbit α-like globin gene cluster. J Biol Chem 262: 5414–5421.
Demuth JP, De Bie T, Stajich JE, Cristianini N, Hahn MW (2006). Evolution of mammalian gene families. PLoS ONE 1: e85.
Excoffier L, Laval G, Schneider S (2005). Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinform Online 1: 47–50.
Excoffier L, Slatkin M (1995). Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol 12: 921–927.
Ferrand N, Azevedo M, Mougel F (2000). A diallelic short tandem repeat, (CCCCG) 4 or 5, located in intron 1 of rabbit α-globin gene. Anim Genet 31: 74.
Ferrand N, Branco M (2007). The evolutionary history of the European rabbit (Oryctolagus cuniculus): major patterns of population differentiation and geographic expansion inferred from protein polymorphism.. In: Weiss S, Ferrand N. (eds). Phylogeography of European Refugia. Springer: Netherlands. pp 207–235.
Fonseca A (2006) Análise da origem e dispersão das populações de coelho-bravo (Oryctolagus cuniculus) nos Arquipélagos dos Açores, Madeira e Canárias, através da utilização de marcadores genéticos nucleares e de DNA mitocondrial. PhD Thesis, University of Açores.
Geraldes A, Carneiro M, Delibes-Mateos M, Villafuerte R, Nachman MW, Ferrand N (2008). Reduced introgression of the Y chromosome between subspecies of the European rabbit (Oryctolagus cuniculus) in the Iberian peninsula. Mol Ecol 17: 4489–4499.
Geraldes A, Ferrand N, Nachman MW (2006). Contrasting patterns of introgression at X-linked loci across the hybrid zone between subspecies of the European rabbit (Oryctolagus cuniculus). Genetics 173: 919–933.
Geraldes A, Rogel-Gaillard C, Ferrand N (2005). High levels of nucleotide diversity in the European rabbit (Oryctolagus cuniculus) SRY gene. Anim Genet 36: 349–351.
Godinho R, Mendonca B, Crespo EG, Ferrand N (2006). Genealogy of the nuclear beta-fibrinogen locus in a highly structured lizard species: comparison with mtDNA and evidence for intragenic recombination in the hybrid zone. Heredity 96: 4454–4463.
Hahn MW, Demuth JP, Han S-G (2007). Accelerated rate of gene gain and loss in primates. Genetics 177: 1941–1949.
Hardison R, Krane D, Vandenbergh D, Cheng JF, Mansberger J, Taddie J et al. (1991). Sequence and comparative analysis of the rabbit α-like globin gene cluster reveals a rapid mode of evolution in a G+C-rich region of mammalian genomes. J Mol Biol 222: 233–249.
Higgs DR (2001). Molecular mechanisms of α-thalassemia.. In: Steinberg MH, Forget BG, Higgs DR, Nagel RL (eds). Disorders of Hemoglobin: Genetics, Pathophysiology and Clinical management. Cambridge University Press: Cambridge. pp 405–430.
Hoffman SMG, Brown WM (1995). The molecular mechanism underlying the ‘rare allele phenomenon’ in a subspecific hybrid zone of the California field mouse, Peromyscus californicus. J Mol Evol 41: 1165–1169.
Hoffmann FG, Gorr TA, Storz JF, Opazo JC (2010). Lineage-specific patterns of functional diversification in the α- and β-globin gene families of tetrapod vertebrates. Mol Biol Evol 27: 1126–1138.
Hoffmann FG, Opazo JC, Storz JF (2008a). Rapid rates of lineage-specific gene duplication and deletion in the α-globin gene family. Mol Biol Evol 25: 591–602.
Hoffmann FG, Opazo JC, Storz JF (2008b). New genes originated via multiple recombinational pathways in the β-globin gene family of rodents. Mol Biol Evol 25: 2589–2600.
Hudson RR (1987). Estimating the recombination parameter of a finite population model without selection. Genet Res 50: 245–250.
Hudson RR, Kaplan NL (1985). Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111: 147–164.
Librado P, Rozas J (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452.
Nei M (1987). Molecular Evolutionary Genetics. Columbia University Press: New York.
Opazo JC, Hoffmann FG, Storz JF (2008a). Differential loss of embryonic globin genes during the radiation of placental mammals. Proc Natl Acad Sci USA 105: 12950–12955.
Opazo JC, Hoffmann FG, Storz JF (2008b). Genomic evidence for independent origins of beta-like globin genes in monotremes and therian mammals. Proc Natl Acad Sci USA 105: 1590–1595.
Opazo JC, Sloan AM, Campbell KL, Storz JF (2009). Origin and ascendancy of a chimeric fusion gene: the beta/delta-globin gene of paenungulate mammals. Mol Biol Evol 26: 1469–1478.
Runck AM, Moriyama H, Storz JF (2009). Evolution of duplicated β-globin genes and the structural basis of hemoglobin isoform differentiation in Mus. Mol Biol Evol 26: 2521–2532.
Runck AM, Weber RE, Fago A, Storz JF (2010). Evolutionary and functional properties of a two-locus β-globin polymorphism in Indian house mice. Genetics 184: 1121–1131.
Schilthuizen M, Gittenberger E (1994). Parallel evolution of an sAat ‘hybrizyme’ in hybrid zones in Albinaria hippolyti (Boettger). Heredity 73: 244–248.
Schilthuizen M, Hoekstra RF, Gittenberger E (1999). Selective increase of a rare haplotype in a land snail hybrid zone. Proc R Soc Lond B7 266: 2181–2185.
Schilthuizen M, Hoekstra RF, Gittenberger E (2004). Hybridization, rare alleles and adaptive radiation. Trends Ecol Evol 19: 404–405.
Storz JF, Baze M, Waite JL, Hoffmann FG, Opazo JC, Hayes JP (2007). Complex signatures of selection and gene conversion in the duplicated globin genes of house mice. Genetics 177: 481–500.
Storz JF, Hoffmann FG, Opazo JC, Moriyama H (2008). Adaptive functional divergence among triplicated α-globin genes in rodents. Genetics 178: 1623–1638.
Storz JF, Runck AM, Moriyama H, Weber RE, Fago A (2010). Genetic differences in hemoglobin function between highland and lowland deer mice. J Exp Biol 213: 2565–2574.
Storz JF, Runck AM, Sabatino SJ, Kelly JK, Ferrand N, Moriyama H et al. (2009). Evolutionary and functional insights into the mechanism underlying high-altitude adaptation of deer mouse hemoglobin. Proc Natl Acad Sci USA 106: 14450–14455.
Tajima F (1983). Evolutionary relationship of DNA sequences in finite populations. Genetics 105: 437–460.
Watterson GA (1975). On the number of segregating sites. Theor Popul Biol 7: 256–276.
Woodruff DS (1989). Genetic anomalies associated with Cerion hybrid zones: the origin and maintenance of new electrophoretic variants called hybrizymes. Biol J Linn Soc Lond 36: 281–294.
Zimmer EA, Martin SL, Beverley SM, Kan YW, Wilson AC (1980). Rapid duplication and loss of genes coding for the alpha chains of hemoglobin. Proc Natl Acad Sci USA 77: 2158–2162.
Acknowledgements
We thank J Melo-Ferreira, M Carneiro, M Schilthuizen and two anonymous reviewers for helpful comments on the manuscript. Part of the work was done in the laboratories of the Centro de Malária e Doenças Tropicais (CMDT-LA) and for that we are grateful to Prof Henrique Silveira. We also wish to thank Bruno Monteiro for help in the lab. This work was supported by Fundação para a Ciência e a Tecnologia (PhD grant SFRH/BD/1409/2000 to RC and project grant POCTI/BSE/40280/2001). JFS acknowledges grant support from the National Science Foundation (DEB-0614342 and IOS0949931) and the National Institutes of Health/National Heart, Lung and Blood Institute (R01 HL087216 and HL087216-S1).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Heredity website
Rights and permissions
About this article
Cite this article
Campos, R., Storz, J. & Ferrand, N. Copy number polymorphism in the α-globin gene cluster of European rabbit (Oryctolagus cuniculus). Heredity 108, 531–536 (2012). https://doi.org/10.1038/hdy.2011.118
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2011.118