Abstract
Major histocompatibility complex (MHC) class I-linked microsatellite data and parental assignment data for a group of wild brown trout (Salmo trutta L.) provide evidence of closer spatial aggregation among fry sharing greater numbers of MHC class I alleles under natural conditions. This result confirms predictions from laboratory experiments demonstrating a hierarchical preference for association of fry sharing MHC alleles. Full-siblings emerge from the same nest (redd), and a passive kin association pattern arising from limited dispersal from the nest (redd effect) would predict that all such pairs would have a similar distribution. However, this study demonstrates a strong, significant trend for reduced distance between pairs of full-sibling fry sharing more MHC class I alleles reflecting their closer aggregation (no alleles shared, 311.5±(s.e.)21.03m; one allele shared, 222.2±14.49m; two alleles shared, 124.9±23.88m; P<0.0001). A significant trend for closer aggregation among fry sharing more MHC class I alleles was also observed in fry pairs, which were known to have different mothers and were otherwise unrelated (ML-r=0) (no alleles: 457.6±3.58m; one allele (422.4±3.86 m); two alleles (381.7±10.72 m); P<0.0001). These pairs are expected to have emerged from different redds and a passive association would then be unlikely. These data suggest that sharing MHC class I alleles has a role in maintaining kin association among full-siblings after emergence. This study demonstrates a pattern consistent with MHC-mediated kin association in the wild for the first time.
Similar content being viewed by others
Introduction
The kin selection theory suggests that an individual may improve its inclusive fitness by behaving in a manner that enhances the reproductive success of relatives (Hamilton, 1964; Keller and Ross, 1998). A prediction of the theory is that the level of behavioral interactions between individual organisms should be influenced by the degree of relatedness between the individuals. The magnitude of the interaction can be represented by Hamilton's rule; rb−c>0, where r is the coefficient of relatedness, b the benefit in fitness for the recipient and c the fitness cost to the actor. Following Hamilton's rule, the effectiveness of kin-mediated cooperative social behavior should be determined by the capacity of animals to accurately direct such behavior towards bona fide relatives.
Ancillary kin bias may occur in circumstances in which interactions tend to be with related rather than with unrelated individuals (Sherman and Holmes, 1985). This may be an effect of limited dispersal from natal sites or shared microhabitat preferences among siblings (Barnard et al., 1991; Hepper, 1991). This is a form of passive kin association lending itself to altruistic behavior. Other discrimination mechanisms include previous association and phenotype matching (Holmes and Sherman, 1983). Kin recognition by previous association refers to a requirement for a period of familiarization between the discriminator and the recipient, normally before weaning or fledging. Phenotype matching entails the learning of phenotypic cues from self or from kin that allows for kin discrimination in which the target has not been previously encountered. A number of studies have been conducted to identify which of these two theories applied (Waldman, 1981; Grau, 1982; Holmes and Sherman, 1982; O’Hara and Blaustein, 1982; Frederickson and Sackett, 1984; Halpin and Hoffman, 1987). The bulk of evidence supports recognition by previous association (Porter et al., 1981; Dewsbury, 1982; Holmes and Sherman, 1982; Gavish et al., 1984; Boyd and Blaustein, 1985; Halpin and Hoffman, 1987), although there is also evidence of phenotype matching (Blaustein and O’Hara, 1981, 1982; Grau, 1982).
The nature of cues used in kin recognition is of considerable interest to kin selection. A survey of cues used for kin discrimination found that visual cues were rare and that olfactory or auditory cues were more common (Halpin, 1991). Reaction to these cues can be discrete, identifying that the individual is related to the discriminator. The reaction can also be proportional, in response to the degree of perceived relatedness, and should be context dependent (Harris et al., 2003). In the latter case, it is likely that the number of cues and the number of variants thereof is critical to fine discrimination in the degree of relatedness. Hamilton (1964) hypothesized the existence of a set of ‘recognition alleles’, which entailed the same or closely linked genes to both produce a phenotype that can be recognized by the bearer of the same set of genes and to cause the individual concerned to act altruistically to other bearers of the gene. The theory was popularized by Dawkins (1976) who termed it the ‘green-beard effect’. A ‘green-beard’ gene produces a perceptible trait (for example, the green beard), allows recognition of other individuals with the same gene and also leads to preferential treatment towards other individuals sharing the gene. The gene provides for direct recognition of copies of itself without concern for average relatedness. These genes can act as ‘selfish genes’—genes that are selected for without consideration of their broader adaptive value to the individual. For instance, it is feasible that otherwise unrelated individuals who share ‘green-beard’ genes could recognize each other as ‘kin’ and that this would affect their behavior towards one another. These ‘green-beard’ genes were first presented as hypothetical mediators of altruism and kin selection. ‘Green-beard’ genes have now been identified in the red imported fire ant (Solenopsis invicta) (Keller and Ross, 1998) and in yeast (Saccharomyces cerevisiae) (Smukalla et al., 2008).
The underlying gene(s) that could facilitate cues that can be used to assess relatedness need to be highly variable. Attention was soon drawn to the major histocompatibility complex (MHC) gene family, which is critical to determining self from non-self in immune system function in vertebrates. Classical MHC class Ia molecules are found on the surface of all nucleated cells of the body. These are composed of a heavy and light chain encoded by polymorphic MHC class Ia genes and the invariant β2-microglobulin gene. MHC class II genes, in contrast, are only expressed in a reduced set of cells, for example, antigen-presenting cells such as dendritic cells, B cells and macrophages. MHC class I and II function in the presentation of self and non-self peptides derived from endogenously (that is, mutated, misfolded or viral) and exogenously (for example, bacterial or macroparasitic) derived proteins, to cytotoxic T lymphocytes or helper T cells, respectively.
Major histocompatibility complex diversity is considered crucial for the ability of populations to resist disease challenges (O’Brien and Evermann, 1988; Muirhead, 2001; Bernatchez and Landry, 2003; Kurtz et al., 2004). MHC may also be used as ‘recognition alleles’ for helping to identify related from unrelated individuals (Hamilton, 1964; Dawkins, 1976; Manning et al., 1992). This process may be used in discriminatory behavior in kin and sexual selection.
The ability of organisms to detect MHC types provides a mechanism for differential behavior between MHC types. Identification of olfactory-based kin recognition mediated by MHC was first demonstrated in rodents (Yamazaki et al., 1976; Brown et al., 1987). Humans also seem to have the capacity to discriminate among conspecifics based on MHC (Wedekind et al., 1995; Ober et al., 1997; Chaix et al., 2008). Olfaction seems to be important in behavior in freshwater fish (Burnard et al., 2008). There has also been extensive evidence for MHC-based mate selection (Reusch et al., 2001; Milinski et al., 2005, 2010; Boehm and Zufall, 2006; Forsberg et al., 2007; Consuegra and Garcia de, 2008; Neff et al., 2008; Eizaguirre et al., 2009; Miller et al., 2009; Turner et al., 2009; Agbali et al., 2010) and kin discrimination (Olsen et al., 1998, 2002; Rajakaruna et al., 2006; Gerlach et al., 2008) in fish. However, kin discrimination has not been demonstrated in the wild to date.
Major histocompatibility complex class I (UBA) and class II (DAA/DAB) have only one expressed locus each in salmonids and these loci are not linked (Shum et al., 2001; Aoyagi et al., 2002; Grimholt et al., 2002; Stet et al., 2002). In this respect, these genes are not a ‘complex’, as in other vertebrates, and are referred to as ‘major histocompatibility’ (MH) genes in salmonids. However, we will use the more commonly used acronym ‘MHC’ to refer to them in this paper. In the laboratory setting, salmonids have been shown to prefer to associate with individuals with which they share more MHC alleles (Olsen et al., 1998, 2002; Rajakaruna et al., 2006). We hypothesize that in the wild, kin-discriminatory behavior should manifest itself as spatial association based on the hierarchical sharing of MHC alleles.
The breeding behavior of salmonids, which involves the construction of nests (redds) in gravel in freshwater, suggests that young salmonids are likely to be found in relatively highly related groups (Hansen et al., 1997; Carlsson et al., 1999; Vera et al., 2010). This has been previously considered a purely passive association because of limited dispersal from natal redds (Elliot, 1987; Hansen et al., 1997), which is driven by stochastic factors (Gowan et al., 1994). However, trout (Salmo trutta L.) were found to demonstrate kin-biased distribution in both juveniles and older trout (Carlsson et al., 2003). These authors suggested that the persistence of kin association in older age groups implied active association, although this could not be proven with the data they had available to them. However, Vera et al. (2010) found no evidence of association among older trout in Spain.
We tested the hypothesis of active spatial association based on the hierarchical sharing of MHC alleles in wild populations of trout fry, separate from any passive association caused by a redd effect.
Materials and methods
We examined spatial association of trout fry based on the sharing of MHC class I-linked microsatellite marker alleles over a ∼1.2 km stretch of the Srahrevagh River.
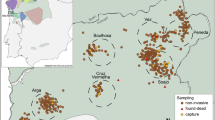
The Srahrevagh is an oligotrophic river (Dineen et al., 2007) in Co. Mayo, Ireland, which has traps operated by the Marine Institute of Ireland, which allow fish moving upstream and downstream to be counted and sampled (Figure 1). On 26 August 2002, 242 trout fry were obtained by electrofishing the trapped stretch of the Srahrevagh River. The river was divided into sections, which were ∼200 m apart (range 110–230 m). The start and end of each section were mapped with GPS, and marked with stakes. Sections were fished once in an upstream direction, by one person with a backpack electrofisher, followed by one person with a net. All stunned trout were netted and held in buckets. Each section was fished up to a natural barrier at the top of the section (for example, small fall) to minimize fish movement out of the section. Distances between sites were considered the distance between the top of each section. Fry that were found in the same site were given a distance of zero. Finer geographic sampling may have improved resolution power, but this was not possible in the context of the current study. The distance data were tested for normality using a Kolmogorv–Smirnov test. Tissue samples were stored in absolute ethanol.
DNA extractions were conducted by dissecting small pieces of tissue (1–5 μg) from the samples and added to 0.5 ml tubes containing 300 μl of 10% (w/v) Chelex solution (MB Chelex-100 resin; Bio-Rad Laboratories, Hercules, CA, USA). The mixture was heated at 99 °C for 1 h. Samples were centrifuged at 3000 r.p.m. for 3 min and then stored at −20 °C.
All 242 individuals were screened at the selectively neutral microsatellite loci Str73 (Estoup et al., 1993), Ssa85 and Ssa197 (O’Reilly et al., 1996), Ssa2216 (Paterson et al., 2004), SsOsl417 and SsOsl85 (Slettan et al., 1995), F43 (Sanchez et al., 1996) and Str543 (Presa and Guyomard, 1996), and a microsatellite locus embedded in the untranslated 3′ end of MHC class I locus (Satr-UBA) (Grimholt et al., 2002). This dinucleotide microsatellite marker is located in the 3′ untranslated region of the MHC class I locus and has been demonstrated to be tightly linked to class I locus in Atlantic salmon (Salmo salar L.) (Grimholt et al., 2002). It has been successfully used previously in studies in salmon (de Eyto et al., 2007; Consuegra et al., 2011) and trout (Coughlan et al., 2006; Hansen et al., 2007). It was not feasible to perform Satr-UBA sequencing of the fry as Satr-UBA has a 14 000 bp intron between exons II and III, which code for the α1 and α1 domains. This means that cDNA must first be derived from RNA before PCR and sequencing could be conducted. The tissue samples available were stored in ethanol meaning that this approach could not be undertaken.
Satr-UBA sequencing of 30 Srahrevagh trout identified 21 alleles, in which the linked microsatellite locus had only 13 alleles (P<0.0001) (Coughlan et al., 2006). The marker itself is not involved in any putative kin recognition. Allele sharing at the marker is a reasonable and conservative measure of allele sharing at Satr-UBA, as demonstrated in simulation tests below.
PCR amplifications were carried out under the following conditions in a 10 μl reaction volume: 95 °C 3 min (95 °C for 30 s, 56 °C for 30 s and 72 °C for 30 s) × 30 cycles. Alleles were resolved on 18 or 25 cm 6% polyacrylamide gels, using a Li-Cor 4200 DNA sequencer (Li-Cor, Lincoln, NE, USA). Allele sizes were determined by reference to a 50–350 bp size ladder and locus-specific allele size standards. These allele size standards were constructed in the laboratory using the full complement of allele sizes observed in pilot studies, to enable consistent scoring among batches of individuals screened for each locus. When initial genotyping was unclear because of gel electrophoresis problems or weak amplification (∼85 genotypes out of a total of 2178), trout fry samples were re-extracted and re-screened. Large allele dropout was identified as an occasional problem but large alleles could usually be reliably scored after re-screening. The estimated error rate was ⩽0.5% of composite genotypes per individual (Coughlan et al., 2006).
Statistical analysis
A rarefaction analysis (Altmann et al., 1996; de Ruiter and Geffen, 1998) was conducted using the web-based program RE-RAT (Schwacke and Rosel, 2005). This tested the robustness of relatedness estimates (Lynch and Ritland, 1999) using the eight neutral microsatellite loci. Initially, pairwise matrices of relatedness values were calculated from a single random locus. After 1000 simulations, the procedure provided mean differences in relatedness values with s.d. as each additional locus was added.
Each unique pair of fry was classified according to whether zero, one or two MHC class I marker alleles were shared. The eight neutral microsatellite markers were used to determine relatedness between fry pairs. The program ML-RELATE (Kalinowski et al., 2006) was used for maximum likelihood relatedness value (ML-r) estimation between pairs of individuals. Full-sibling pairs were identified as those showing ML-r⩾0.5. These full-sibling pairs are expected to have emerged from the same nest (redd).
Parental assignment data were available from sampling adults moving upstream to spawn in the autumn of the previous year (2001). Parental assignment data were then used to identify those pairs of fry (among assigned fry) that did not share a mother. These fry were expected not to have emerged from the same redd. Fry pairs in this sub-group with ML-r of 0 were then identified and the remaining pairs (Ml-r>0) removed from the analysis. These fry were then also unlikely to be paternal half siblings. This was our ‘unrelated’ group. It should be noted that the large number of uncollected parents in the parental assignment data available meant very few full-siblings were identified, necessitating using the relatedness-based approach to identify full-siblings instead.
Separate analyses were then conducted for the full-sibling and unrelated groups. The mean distances between pairs sharing different numbers of MHC alleles were calculated, Kruskal–Wallis tests and, subsequent, pairwise Mann–Whitney tests between groups conducted. However, because fish separated by very large distances may be equally unlikely to interact with each other, we wanted to account for this through a second analysis in which pairs were binned by distance to ‘within section’, ‘neighboring section’ (one section away) and ‘distant sections’ (more than one section away). The number of pairs sharing zero, one and two MHC alleles within each distance category for the observed data were incorporated into a contingency table. A bootstrap procedure was then implemented in Python scripts wherein each pair of trout fry had assigned to it a random value from the observed distribution of MHC marker allele shared values, with replacement. The proportion of 10 000 bootstrap values which exceeded or were less than the observed value in each cell of the contingency table were used to determine whether observed values differed significantly from random expectations. The bootstrap procedure was preferable to a parametric approach such as the χ2 test in that it provided for any imbalance in the experimental design which might result in unequal variances in the different groups.
Simulation study of microsatellite–UBA correlation for full-sibling groups
We tested the validity of using the microsatellite marker as a proxy for UBA allele sharing by simulating full-sibling families based on the available microsatellite data from this study and the Satr-UBA data from the same river (Coughlan et al., 2006). The Satr-UBA data were generated from 27 resident brown trout adults selected to represent all microsatellite alleles identified in a sample of 107 fish in 2004 from the Srahrevagh river, Co. Mayo. For each microsatellite allele, sets of UBA alleles observed in association with each microsatellite allele were compiled. The 110 and 136 bp microsatellite alleles (136 bp was most common) were always found with a particular UBA allele, Satr-UBA*1701 and Satr-UBA*1201, respectively.
At each iteration of the procedure, an independent full-sibling data set was simulated and analyzed. A set of parental genotypes were then simulated based on the observed MHC microsatellite data for the Srahrevagh 2002 trout fry sample. The numbers of males and females were independently selected, a random number between 40 and 50 in both cases. Each parent has a microsatellite genotype drawn at random from the observed microsatellite allele frequency distribution available for the Srahrevagh population. For each microsatellite allele in the parent's genotype, a random UBA allele is drawn from the observed set of UBA alleles associated that particular microsatellite allele (Coughlan et al., 2006). Each parent then has a pair of MHC ‘chromosomes’ with a particular combination of a UBA and a microsatellite allele in each.
A maximum family size was selected at random between 80 and 100. We then created 125 full-sibling families by randomly ‘mating’ two simulated parents. The number of fry in each family was randomly selected between 10 and the maximum family size for each iteration. For each fry simulated, a random MHC ‘chromosome’ is inherited from the father and one from the mother.
After generation of every full-sibling family, the MHC ‘chromosome’ genotype of each fry in the family was compared with that of every other fry in the family. In each pairwise comparison, the number of microsatellite alleles shared among the pair was compared with the number of UBA alleles shared among the pair. If the number is the same, a ‘correct’ score is logged under the appropriate category of microsatellite alleles shared (zero, one or two). If the number of UBA alleles shared differs, an ‘incorrect’ score was logged under the appropriate category. At the end of each iteration, the proportion of ‘correct’ to ‘incorrect’ scores were calculated for the zero, one and two alleles shared categories. The procedure was repeated across 1000 bootstraps. These values were then summarized across the 1000 iterations of the simulation study. The simulations mirrored the approach taken in our key analysis of full-sibling groups identified in the real data from relatedness values. The simulation results gave us an idea of how well the microsatellite allele sharing patterns fitted those at UBA within the full-sibling groups simulated from the Srahrevagh data. This also gave us an estimation of how well we might expect the microsatellite marker to perform for identifying UBA allele sharing in our real data.
Results
Distance data among all pairs of fry were found to be normally distributed (Kolmogorv–Smirnov test, N=29 161, Z=22.187, P=0.000). The rarefaction analysis demonstrated that resolution power with any seven of the eight neutral loci provided robust relatedness estimation. The total numbers of alleles identified in the 242 fry for each locus were as follows: Str73 (total alleles: NA=3, expected heterozygosity: HE=0.580), Ssa85 (NA=5, HE=0.730), Ssa197 (NA=8, HE=0.704), Ssa2216 (NA=6, HE=0.673), SsOsl417 (NA=21, HE=0.772) and SsOsl85 (NA=18, HE=0.883), F43 (NA=13, HE=0.823) and Str543 (NA=12, HE=0.809). The MH class I-linked microsatellite had 11 alleles and an expected heterozygosity of 0.825.
Full-sibling analysis
A total of 749 full-sibling pairs were identified. Both individuals in each of these full-sibling pairs would have emerged from the same nest (redd). A passive kin association pattern, arising from limited dispersal from the nest (redd effect) would predict that all such pairs would have a similar distribution. However, this study demonstrates a strong, significant and hierarchical trend for closer aggregation among full-sibling fry sharing more MHC class I marker alleles, as shown by a reducing mean distance between pairs sharing more alleles (no alleles shared: mean=311.5±21.03 m, median=170 m, mean ranks=422.02, n=248; one allele shared: 222.2±14.49 m, median=150 m, mean ranks=362.01, n=407; two alleles shared: 124.9±23.88 m, median=0 m, mean ranks=295.80, n=86; Kruskal–Wallis test (H (df=2)=27.537, df=2, P<0.0001, Figure 2). Details of all tests (including pairwise Mann–Whitney U-tests) can be found in Table 1.
Box plot of full-sibling fry distribution data. Outliers (values that are between 1.5 and 3 times the interquartile range) are represented by circles, whereas extreme outliers (values that are over 3 times the interquartile range) are marked with asterisks. The whiskers represent the minimum and maximum values which are not outliers.
Full-sibling pairs that shared two MHC alleles were more frequent than expected within a section (Bootstrap test, P=0.0029), not significantly different from expectation at neighboring sections (P=0.1787), and less frequent than expected at distant sections (P=0.0001) (Table 2). Those that shared one MHC allele were more frequent than expected within a section (P=0.0262), but not significantly different from expectation at neighboring sections (P=0.1699) or distant sections (P=0.3276). Full-sibling pairs sharing no MHC alleles were significantly fewer than expected within section (P=0.0001), not significantly different from expectation at neighboring sections (P=0.4679), and more frequent than expected at distant sections (P=0.0000). Full-sibling pairs which shared two MHC alleles were rarely found more than two sections away from each other, whereas full-sibling pairs that shared no MHC alleles were unusually common in these ‘distant sections’ (see Table 2).
Therefore, full-sibling fry that emerged from the same redd not only tended to be found closer to fry with which they share more MHC alleles, but to have dispersed less than those sharing fewer alleles. Full-siblings who share two MHC alleles demonstrate a particularly marked tendency for association, whereas those that share no MHC alleles show much poorer association. This suggests that sharing MHC class I alleles has a role in maintaining kin association among these full-siblings after emergence.
Unrelated fry analysis
Parental assignment data were available from genotyping adults sampled moving upstream to spawn in the previous year. This parental assignment data were based, primarily, on complete exclusion as the only situation in which successful allocation of offspring is difficult to question (Jones and Ardren, 2003). Offspring of sampled males were found over a mean range of ∼250 m, whereas those of sampled females were found over a mean range of ∼400 m. The ‘unrelated’ group consisted of 15 006 pairs. We know that these pairs did not emerge from the same redd as they do not share a mother. Redd effect is the most likely source of passive association. A paternal effect on fry spawned in adjacent sections by the same father may be possible, but our exclusion of fry pairs with Ml-r>0 should help exclude half siblings.
A significant trend for closer aggregation among fry sharing more MHC class I alleles was also observed in these unrelated pairs (zero alleles, mean=457.6±3.58 m, median=430 m, mean ranks=7677.22; one allele, mean=422.4±3.86 m, median=350 m, mean ranks=7341.41; two alleles, mean=381.7±10.72 m, median=345 m, mean ranks=7198.54; Kruskal–Wallis test, H (df=2)=25.503, P<0.0001, see Figure 3). However, no significant difference was found in the pairwise Mann–Whitney test for unrelated fry sharing one allele and two alleles (U=2 551 245, Z=−0.851, P=0.395).
Unrelated pairs sharing two MHC alleles (P=0.0197) and one MHC allele (P=0.0001) were significantly more likely to be found in the same section (Table 2). Conversely, those sharing no alleles were significantly less likely to be found in the same section (P=0.0000). No significant pattern was observed for fry sharing different numbers of MHC alleles in neighboring sections (∼100 m) (Table 2). Fry sharing one MHC allele were significantly less likely to be found in distant sections (⩾200 m) (P=0.0041) but, unexpectedly, there was no such significant pattern for fry sharing two alleles (P=0.0722). Fry sharing no MHC alleles were significantly more common in distant sections (P=0.0009). Overall, these data suggest that MHC also significantly affects the distribution of these unrelated brown trout fry. This then also appears to suggest an active association based on the sharing of MHC class I alleles, given a passive association is unlikely.
Simulation study of microsatellite–UBA correlation for full-sibling groups
Our simulation of full-sibling groups, based on the available microsatellite and UBA data, appeared to support the efficacy of the microsatellite marker in identifying allele sharing at the UBA locus. More than 1000 bootstrap iterations of the simulation, full-siblings that shared no microsatellite also shared no UBA alleles 90.0% of the time (s.e. 0.24%), those which shared one microsatellite allele shared one UBA allele 84.9% of the time (s.e. 0.14%) and those which shared two microsatellite alleles also shared two UBA alleles 81.7% of the time (s.e. 0.27%). This implies that, when applied to full-sibling groups, the microsatellite marker is a good indicator of UBA allele sharing among pairs of fish. The marker predicts pairs sharing two alleles somewhat less accurately. If there is any bias caused by this, there is no obvious reason why this would lead us to identify UBA-mediated kin association in which there is none. It seems more likely that inaccurate inclusion of pairs sharing two marker alleles which do not share two UBA alleles, for instance, would weaken the power of our analysis to identify an underlying trend of association based on UBA allele sharing. This could occur wherein pairs with no behavioral preference for one another and which might be expected to be found a random distance apart, are treated the same as those that share two UBA alleles and may demonstrate a strong behavioral preference for each other.
Discussion
To our knowledge, this is the first time kin association based on the sharing of MHC alleles has been demonstrated in the wild. Previous laboratory studies had found evidence for this hierarchical preference for fry sharing more MHC alleles (Rajakaruna et al., 2006), but that study had examined MHC class II. The authors did suggest that their data implied that another locus, perhaps MHC class I, was also affecting behavioral preferences. Our data would seem to suggest that MHC class I is a factor in kin recognition in these fish. Unrelated fry also showed signs of greater association based on the sharing of MHC alleles. This could also be a consequence of the use of MHC in kin recognition. Unrelated salmonids were found to prefer non-kin sharing two MHC class II alleles over non-kin with which they did not share alleles in the laboratory studies (Rajakaruna et al., 2006). Evidence for an influence of a MHC-based kin recognition system in unrelated fry, both in their behavior and in their distribution in the wild, may be indicative of a form of ‘green-beard effect’ (Dawkins, 1976) occurring in salmonids fish.
Laboratory studies of kin-mediated cooperative social behavior in salmonids (Quinn and Busack, 1985; Quinn and Hara, 1986; Olsen, 1989; Brown and Brown, 1992, 1993, 1996; Hedenskog et al., 2002) would suggest that the benefits of active kin association in these fry would be in the acquisition and maintenance of territories after emergence from the redd. Trout that do not succeed in maintaining a territory are assumed to die, with >90% mortality during this critical period (Elliot, 1994). This constitutes a powerful selective pressure.
Altruistic behavior will be most effective when directed exclusively at full-sibling relatives and less so when directed against half-siblings (Hamilton, 1964). Thus, the ability for trout fry to discriminate between full-sibling relatives and half-sibling relatives would be valuable. Any kin recognition system will only be as effective as the resolution power achievable by the genetic loci and/or phenotypic cues used. In this respect, the use of the highly polymorphic MHC loci is particularly valuable.
The resolution power of highly polymorphic loci such as MHC depends not just on the number of alleles within the population at large, but on the relative frequencies of each allele. The number of alleles may vary from one population to the next with varying degrees of skew in the frequencies of each allele. Individuals that share common alleles may identify a greater proportion of conspecifics as related when they are not, with fitness consequences. Conversely, individuals that encounter each other and share rare alleles may be more likely to be bona fide relatives.
A ‘rare allele advantage’ for kin recognition systems has been suggested by Grafen (1990). In populations that exhibit very skewed allele frequencies or low polymorphism in general (perhaps due to an epizootic or population bottleneck), the resolution ability of kin recognition is greatly reduced for any individual possessing one or more common MHC alleles. In contrast, individuals with rarer alleles enjoy a relative selective advantage arising from very effective cooperative behaviors. Such an advantage is inversely proportional to the frequency of the bearer's allele(s) in the population and may continue to accrue over several generations. A prediction of this theory would be that accurate kin recognition breaks down wherein MHC variation is reduced in a population. Empirical evidence of kin association, using a genome-wide measure such as relatedness, may not be found wherein kin recognition breaks down. However, if kin recognition is based on additional genes, then kin recognition, although less accurate, could still operate after a bottleneck event which reduced MHC variability. This may provide an additional explanation for the retention of high levels of polymorphism at MHC loci due to balancing selection. Heterozygosity and the presence of rare MHC alleles in offspring genotypes may be beneficial to survival even in the absence of direct disease challenge.
References
Agbali M, Reichard M, Bryjova A, Bryja J, Smith C (2010). Mate choice for nonadditive genetic benefits correlate with MHC dissimilarity in the rose bitterling (Rhodeus ocellatus). Evolution 64: 1683–1696.
Altmann J, Alberts SC, Haines SA, Dubach J, Muruthi P, Coote T et al. (1996). Behavior predicts genes structure in a wild primate group. Proc Natl Acad Sci USA 93: 5797–5801.
Aoyagi K, Dijkstra JM, Xia C, Denda I, Ototake M, Hashimoto K et al. (2002). Classical MHC class I genes composed of highly divergent sequence lineages share a single locus in rainbow trout (Oncorhynchus mykiss). J Immunol 168: 260–273.
Barnard CJ, Hurst JL, Aldhous P (1991). Of mice and kin: the functional significance of kin bias in social behaviour. Biol Rev 66: 379–430.
Bernatchez L, Landry C (2003). MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? J Evol Biol 16: 363–377.
Blaustein AR, O’Hara RK (1981). Genetic control for sibling recognition? Nature 290: 246–248.
Blaustein AR, O’Hara RK (1982). Kin recognition in Rana cascadae tadpoles: maternal and paternal effects. Animal Behav 30: 1151–1157.
Boehm T, Zufall F (2006). MHC peptides and the sensory evaluation of genotype. Trends Neurosci 29: 100–107.
Boyd SK, Blaustein AR (1985). Familiarity and inbreeding avoidance in the gray-tailed vole (Microtus canicaudus). J Mammal 66: 348–352.
Brown G, Brown J (1992). Do rainbow trout and Atlantic salmon discriminate kin? Can J Zool 70: 1636–1640.
Brown G, Brown J (1993). Social dynamics in salmonid fishes: do kin make better neighbors? Animal Behav 45: 863–871.
Brown GE, Brown JA (1996). Does kin-biased territorial behavior increase kin-biased foraging in juvenile salmonids? Behav Ecol 7: 24–29.
Brown RE, Singh PB, Roser B (1987). The major histocompatibility complex and the chemosensory recognition of individuality in rats. Physiol Behav 40: 65–73.
Burnard D, Gozlan RE, Griffiths SW (2008). The role of pheromones in freshwater fishes. J Fish Biol 73: 1–16.
Carlsson J, Carlsson JEL, Olsen KH, Hansen MM, Eriksson T, Nilsson J (2003). Kin-biased distribution in brown trout: an effect of redd location or kin recognition? Heredity 92: 53–60.
Carlsson J, Olsen KH, Nilsson J, Overli O, Stabell OB (1999). Microsatellites reveal fine-scale genetic structure in stream-living brown trout. J Fish Biol 55: 1290–1303.
Chaix R, Cao C, Donnelly P (2008). Is mate choice in humans MHC-dependent? PLoS Genet 4: e1000184.
Consuegra S, De EE, Mcginnity P, Stet RJ, Jordan WC (2011). Contrasting responses to selection in class I and class IIalpha major histocompatibility-linked markers in salmon. Heredity 107: 143–154.
Consuegra S, Garcia De LC (2008). MHC-mediated mate choice increases parasite resistance in salmon. Proc Biol Sci 275: 1397–1403.
Coughlan J, Mcginnity P, O’Farrell B, Dillane E, Diserud O, De Eyto E et al. (2006). Temporal variation in an immune response gene (MHC I) in anadromous Salmo trutta in an Irish river before and during aquaculture activities. ICES J Marine Sci 63: 1248–1255.
Dawkins R (1976). The Selfish Gene. Oxford University Press: Oxford.
De Eyto E, Mcginnity P, Consuegra S, Coughlan J, Tufto J, Farrell K et al. (2007). Natural selection acts on Atlantic salmon major histocompatibility (MH) variability in the wild. Proc Biol Sci 274: 861–869.
De Ruiter JR, Geffen E (1998). Relatedness of matrilines, dispersing males and social groups in long-tailed macaques (Macaca fascicularis). Proc Roy Soc London Ser B-Biol Sci 265: 79–87.
Dewsbury DA (1982). Avoidance of incestuous breeding between siblings in two species of Peromyscus mice. Biol Behav 7: 157–168.
Dineen G, Harrison SSC, Giller PS (2007). Growth, production and bioenergetics of brown trout in upland streams with contrasting riparian vegetation. Freshwater Biol 52: 771–783.
Eizaguirre C, Yeates SE, Lenz TL, Kalbe M, Milinski M (2009). MHC-based mate choice combines good genes and maintenance of MHC polymorphism. Mol Ecol 18: 3316–3329.
Elliot JM (1994). Quantitative Ecology and the Brown Trout. Oxford University Press: Oxford.
Elliot J (1987). The distances travelled by downstream-moving trout fry, Salmo trutta, in a lake district stream. Freshwater Biol 17: 491–499.
Estoup A, Presa P, Krieg F, Vaiman D, Guyomard R (1993). (Ct)(N) and (Gt)(N) Microsatellites—A new class of genetic markers for Salmo trutta L brown trout. Heredity 71: 488–496.
Forsberg LA, Dannewitz J, Petersson E, Grahn M (2007). Influence of genetic dissimilarity in the reproductive success and mate choice of brown trout—females fishing for optimal MHC dissimilarity. J Evol Biol 20: 1859–1869.
Frederickson WT, Sackett GP (1984). Kin preferences in primates: relatedness or familiarity? J Comp Psychol 98: 29–34.
Gavish L, Hoffman JE, Getz LL (1984). Sibling recognition in the prairie vole, Microtus ochrogaster. Animal Behav 32: 362–366.
Gerlach G, Hodgins-Davis A, Avolio C, Schunter C (2008). Kin recognition in zebrafish: a 24-hour window for olfactory imprinting. Proc Biol Sci 275: 2165–2170.
Gowan C, Young MK, Fausch KD, Riley SC (1994). Restricted movement in resident stream salmonids: a paradigm lost? Can J Fisheries Aquat Sci 51: 2626–2637.
Grafen A (1990). Do animals really recognize kin? Animal Behav 39: 42–54.
Grau JH (1982). Kin recognition in white-footed deermice (Peromyscus leucopus). Animal Behav 30: 497–505.
Grimholt U, Drablos F, Jorgensen SM, Hoyheim B, Stet RJM (2002). The major histocompatibility class I locus in Atlantic salmon (Salmo salar L): polymorphism, linkage analysis and protein modelling. Immunogenetics 54: 570–581.
Halpin ZT (1991). Kin recognition cues of vertebrates. In: Hepper PG (ed). Cambridge University Press: Cambridge. pp 220–258.
Halpin ZT, Hoffman MD (1987). Sibling recognition in the white-footed mouse, Permoyscus leucopus: association or phenotype matching? Animal Behav 35: 563–570.
Hamilton W (1964). The genetical evolution of social behaviour. J Theor Biol 7: 1–52.
Hansen MM, Nielsen EE, Mensberg K-LD (1997). The problem of sampling families rather than populations: relatedness among individuals in samples of juvenile brown trout (Salmo trutta L.) Mol Ecol 6: 469–474.
Hansen M, Skaala O, Jensen L, Bekkevold D, Mensberg K-L (2007). Gene flow, effective population size and selection at major histocompatibility complex genes: brown trout in the Hardanger Fjord, Norway. Mol Ecol 16: 1413–1425.
Harris RN, Vess TJ, Hammond JI, Lindermuth CJ (2003). Context-dependent kin discrimination in larval four-toed salamanders Hemidactylium scutatum. Herpetologica 59: 164–177.
Hedenskog M, Petersson E, Järvi T (2002). Agonistic behavior and growth in newly emerged brown trout (Salmo trutta L.) of sea-ranched and wild origin. Aggressive Behav 28: 145–153.
Hepper PG (1991). Recognizing kin: ontogeny and classification. In: Hepper PG (ed). Cambridge University Press: Cambridge.
Holmes WG, Sherman PW (1982). The ontogeny of kin recognition in two species of ground squirrels. Am Zool 22: 491–517.
Holmes WG, Sherman PW (1983). Kin recognition in animals. Am Scient 71: 46–55.
Jones AG, Ardren WR (2003). Methods of parentage analysis in natural populations. Mol Ecol 12: 2511–2523.
Kalinowski ST, Wagner AP, Taper ML (2006). ML-RELATE: a computer program for maximum likelihood estimation of relatedness and relationship. Mol Ecol Notes 6: 576–579.
Keller L, Ross KG (1998). Selfish genes: a green beard in the red fire ant. Nature 394: 573–575.
Kurtz J, Kalbe M, Aeschlimann PB, Haberli MA, Wegner KM, Reusch TB et al. (2004). Major histocompatibility complex diversity influences parasite resistance and innate immunity in sticklebacks. Proc Biol Sci 271: 197–204.
Lynch M, Ritland K (1999). Estimation of pairwise relatedness with molecular markers. Genetics 152: 1753–1766.
Manning CJ, Wakeland EK, Potts WK (1992). Communal nesting patterns in mice implicate MHC genes in kin recognition. Nature 360: 581–583.
Milinski M, Griffiths S, Wegner KM, Reusch TB, Haas-Assenbaum A, Boehm T (2005). Mate choice decisions of stickleback females predictably modified by MHC peptide ligands. Proc Natl Acad Sci USA 102: 4414–4418.
Milinski M, Griffiths SW, Reusch TB, Boehm T (2010). Costly major histocompatibility complex signals produced only by reproductively active males, but not females, must be validated by a ‘maleness signal’ in three-spined sticklebacks. Proc Biol Sci 277: 391–398.
Miller HC, Moore JA, Nelson NJ, Daugherty CH (2009). Influence of major histocompatibility complex genotype on mating success in a free-ranging reptile population. Proc Biol Sci 276: 1695–1704.
Muirhead CA (2001). Consequences of population structure on genes under balancing selection. Evolution 55: 1532–1541.
Neff B, Garner S, Heath J, Heath D (2008). The MHC and non-random mating in a captive population of Chinook salmon. Heredity 101: 175–185.
O’Brien SJ, Evermann JF (1988). Interactive influence of infectious disease and genetic diversity in natural populations. Trends Ecol Evol 3: 254–259.
O’Hara RK, Blaustein AR (1982). Kin preference behavior in Bufo boreas tadpoles. Behav Ecol Sociobiol 11: 43–49.
O’Reilly PT, Hamilton LC, Mcconnell SK, Wright JM (1996). Rapid analysis of genetic variation in Atlantic salmon (Salmo salar) by PCR multiplexing of dinucleotide and tetranucleotide microsatellites. Can J Fisheries Aquat Sci 53: 2292–2298.
Ober C, Weitkamp L, Cox N, Dytch H, Kostyu D, Elias S (1997). HLA and mate choice in humans. Genetics 61: 497–504.
Olsen KH, Grahn M, Lohm J (2002). Influence of MHC on sibling discrimination in Arctic charr, Salvelinus alpinus L. J Chem Ecol 28: 783–795.
Olsen KH, Grahn M, Lohm J, Langefors A (1998). MHC and kin discrimination in juvenile Arctic charr, Salvelinus alpinus L. Animal Behav 56: 319–327.
Olsen KH (1989). Sibling recognition in juvenile Arctic charr, Salvelinus alpinus (L.) J Fish Biol 34: 571–581.
Paterson S, Piertney SB, Knox D, Gilbey J, Verspoor E (2004). Characterization and PCR multiplexing of novel highly variable tetranucleotide Atlantic salmon (Salmo salar L.) microsatellites. Mol Ecol Notes 4: 160–162.
Porter RH, Tepper VJ, White DM (1981). Experiential influences on the development of huddling preferences and sibling recognition in spiny mice. Devel Psychobiol 14: 375–382.
Presa P, Guyomard R (1996). Conservation of microsatellites in three species of salmonids. J Fish Biol 49: 1326–1329.
Quinn T, Busack C (1985). Chemosensory recognition of siblings in juvenile coho salmon (Oncorhynchus kisutch). Animal Behav 33: 51–56.
Quinn T, Hara T (1986). Sibling recognition and olfactory sensitivity in juvenile coho salmon (Oncorhynchus kisutch). Can J Zoo 64: 921–925.
Rajakaruna J, Brown A, Kaukinen K, Miller K (2006). Major histocompatibility complex and kin discrimination in Atlantic salmon and brook trout. Mol Ecol 15: 4569–4575.
Reusch TB, Haberli MA, Aeschlimann PB, Milinski M (2001). Female sticklebacks count alleles in a strategy of sexual selection explaining MHC polymorphism. Nature 414: 300–302.
Sanchez JA, Clabby C, Ramos D, Blanco G, Flavin F, Vazquez E et al. (1996). Protein and microsatellite single locus variability in Salmo salar L. (Atlantic salmon). Heredity 77: 423–432.
Schwacke L, Rosel P (2005). Relatedness Estimation and Rarefaction Analysis Tool (RE-RAT). http://people.musc.edu/∼schwaclh/.Ref Type: Electronic Citation.
Sherman PW, Holmes WG (1985). Kin recognition: issues and evidence. Forstschr Zool 31: 437–460.
Shum BP, Guethlein L, Flodin LR, Adkison MA, Hedrick RP, Nehring RB et al. (2001). Modes of salmonid MHC class I and II evolution differ from the primate paradigm. J Immunol 166: 3297–3308.
Slettan A, Olsaker I, Lie O (1995). Atlantic salmon, Salmo salar, microsatellites at the Ssosl25, Ssosl85, Ssosl311, Ssosl417 loci. Animal Genet 26: 281–282.
Smukalla S, Caldara M, Pochet N, Beauvais A, Guadagnini S, Yan C et al. (2008). FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell 135: 726–737.
Stet RJM, De Vries B, Mudde K, Hermsen T, Van Heerwaarden J, Shum BP et al. (2002). Unique haplotypes of co-segregating major histocompatibility class II A and class II B alleles in Atlantic salmon (Salmo salar) give rise to diverse class II genotypes. Immunogenetics 54: 320–331.
Turner SM, Chaves-Campos J, Dewoody JA (2009). Parental relatedness and major histocompatibility effects on early embryo survivorship in Atlantic salmon. Genetica 137: 99–109.
Vera M, Sanz N, Hansen MM, Almodovar A, Garcia-Marin JL (2010). Population and family structure of brown trout, Salmo trutta, in a Mediterranean stream. Marine Freshwater Res 61: 676–685.
Waldman B (1981). Sibling recognition in toad tadpoles: the role of experience. Z Tierpsychol 56: 341–358.
Wedekind C, Seebeck T, Bettens F, Paepke A (1995). MHC-dependent mate preferences in humans. Proc Roy Soc London Ser B-Biol Sci 260: 755–760.
Yamazaki K, Boyse E, Mike V (1976). Control of mating preferences in mice by genes in the major histocompatibility complex. J Exp Med 144: 1324–1335.
Acknowledgements
HEA PRTLI Cycle 3; Beaufort Marine Research Award: Fish population genetics. Irish Government NDP (2007–2014) administered by the Marine Institute; Inland Fisheries Ireland; Science Foundation Ireland (Microbial Phylogeography 05/FE1/B882); Marine Institute (Newport) field staff; Robert O’Farrell, Richard O’Callaghan, Niamh Kilgallen, Luca Mirimin, Tad Kirakowski and Eugene Finnerty.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
O'Farrell, B., Benzie, J., McGinnity, P. et al. MHC-mediated spatial distribution in brown trout (Salmo trutta) fry. Heredity 108, 403–409 (2012). https://doi.org/10.1038/hdy.2011.87
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2011.87
Keywords
This article is cited by
-
Neural pathways of olfactory kin imprinting and kin recognition in zebrafish
Cell and Tissue Research (2021)